RAMADDA Data Server
The TCO group runs a RAMADDA data server at https://observations.mpimet.mpg.de/repository/ which is used to expose datasets from the internal filesystem at /pool/OBS to the public using OPeNDAP.
Using datasets
In oder to use a published dataset, please navigate to the dataset of interest and copy the link behind the OPeNDAP service on the top right (see image below). You can use this link as if it where a path to a netCDF dataset. I.e. put it into ncdump -h to get an overview or use it in xr.open_dataset if you like to access it in Python.
Note that the most recent pydap version at the time of writing (3.2.2) has a bug, which prevents loading resources from out RAMADDA server. The bug is fixed on the master branch, but it doesn't look like it will be released anywhere soon. Access via the netCDF Python library works fine nonetheless.
Add datasets
In order to add datasets to the RAMMADA server, you'll need to login (ask Hauke or Tobi on how to get an account).
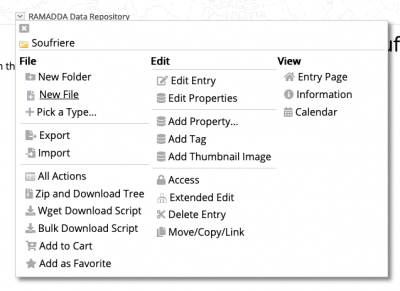
After login, create a new file (the to be published dataset is named a “file”) using the menu:
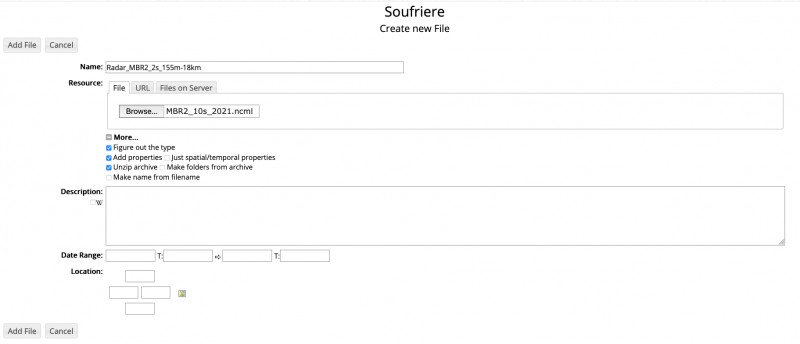
This will direct you to the file creation menu:
You can add a title and description as needed. In order to connect this newly created dataset with data on the /pool/OBS filesystem, you'll have to write an NcML description of the dataset and upload it as a Resource in the RAMADDA file creation menu.
A typical NcML file will look like the following:
<netcdf xmlns="http://www.unidata.ucar.edu/namespaces/netcdf/ncml-2.2"> <aggregation dimName="time" type="joinExisting" recheckEvery="1 day"> <variableAgg name="time"/> <scan location="/srv/data/OBS/BARBADOS_CLOUD_OBSERVATORY/Level_1/K_Radiation" regExp=".*Radiation__Deebles_Point__DownwellingRadiation__1s__2021[0-9]{2}.nc" subfolders="true" /> </aggregation> </netcdf>
This file starts to scan for files in the directory given by location (including subfolders) and selects all files below this directory which match the regular expression given by regExp. If will also rescan for new files every day.
Please note that the /pool/OBS filesystem is reachable via /srv/data/OBS on the RAMADDA server, so you'll have to adjust your paths accordingly.
If you like to check your regular expressions interactively, https://regex101.com is a good place to go and if you want to have some practice on this topic, checkout https://regexcrossword.com 
Fixing Datatypes
OPeNDAP is not able to properly represent 64bit integers. 64bit integers are often used e.g. in time variables, even if that would not be necessary. So sometimes issues with 64bit integers can be resolved just by changing the datatype. Fortunately, NcML has a facility to do so:
<netcdf xmlns="http://www.unidata.ucar.edu/namespaces/netcdf/ncml-2.2"> <aggregation dimName="time" type="joinExisting" recheckEvery="1 day"> <variableAgg name="time"/> <scan location="/srv/data/OBS/BARBADOS_CLOUD_OBSERVATORY/Level_1/B_Reflectivity/Ka-Band/MBR2/10s/" regExp=".*MMCR__MBR2__Spectral_Moments__10s__155m-18km__21.*" suffix=".nc" subfolders="false" /> </aggregation> <variable name="time" type="int"> </variable> </netcdf>