Parallel Analysis with Dask
Ever wondered why your Python script is taking so long to finish?
Often in our daily work as researcher we process large amounts of data. The processing is usually done using utilities like Python or NCL which are, by default, not parallelized. On the other hand models used to produce the data that we analyse are obviously parallelized using libraries like OpenMP or MPI. With the increasing volume of data produced by models it is becoming more and more necessary to parallelise the processing part of data analysis. This can be easily done with Python and the Dask (https://dask.org/) library.
Why Dask?
Dask parallelise the processing of data by (1) dividing the arrays into chunks, which can be defined by the users, and (2) interfacing with the functions of numpy, pandas and scikit-learn which can then operate over these chunks. Every operation is decomposed in many sub-operations which can then be parallelised across different processors. For example in order to compute an average of an array Dask will divide the array into chunks, compute the average over all the individual chunks and then merge everything together. The interesting part of this process is that Dask can be adapted to run either on your laptop or on a cluster where more processors are available. This means that machines that have a limited amount of memory (for example a 'default' macbook used at MPI may have about 8 GB of RAM) can still operate onto array which would not fit into memory since operations are performed over chunks. In other words, you will be able to open files of e.g. 64 GB on a machine that has 8 GB of RAM. For more information check out the official website https://dask.org/.
In this guide we will use Dask to process data of the simulations conducted over Germany in the framework of the HD(CP)2 project. The idea is that we don't want to move/copy any file to compute our final result, which will be just a time series of rain rate. But first of all we need to talk about the configuration.
Configuration
When using Python on Mistral one is advised to use conda to manage the installed packages. This page https://www.dkrz.de/up/systems/mistral/programming/python explains how to install conda and create a virtual environment where all the packages are installed. Assuming that our environment is called my_base to install all the necessary packages one needs to do
source activate my_base conda install -c conda-forge dask xarray dask-jobqueue ipython pandas numpy matplotlib pip install dask-mpi
Note that we used pip to install dask-mpi since this is the only way to correctly link the MPI library needed to launch a job using dask-mpi. Later it will become clear why this is important.
You can use Dask either in ipython or in a jupyter notebook, which I find more convenient. DKRZ created a nice script to launch a jupyter notebook on Mistral which can be found here https://www.dkrz.de/up/systems/mistral/programming/jupyter-notebook. When working with Dask it is advisable to reserve an explicit node on the shared partition so that any potential node for the computation can be easily reached through the network. This can be done specifying the account that need to be used for the allocation of the node:
chmod a+x start-jupyter ./start-jupyter -u m300382 -A bm0682
As documented on the DKRZ website this script will read some configuration from a jupyter_preload file located in your home on Mistral. You should use that file to load your specific virtual environment to make sure that the notebook will have all the packages that you installed. For example
module unload anaconda2 anaconda3 netcdf_c python module load anaconda2/bleeding_edge export PATH=/work/mh0731/m300382/conda_envs/my_base/bin:$PATH source /sw/rhel6-x64/conda/anaconda2-bleeding_edge/bin/activate my_base export PYTHONNOUSERSITE=True
Unfortunately jupyterhub cannot be used at the moment with Dask as the necessary packages cannot be installed in the default environment and a user defined environment cannot be loaded in jupyterhub.
Main difference in the processing workflow
Dask employs lazy arrays. This is a fancy way of saying that array are never loaded into memory if they are not needed for some computation or you don't tell Dask to load. This avoid to fill up the memory of the machine and reduce waiting time for operations. This, however, means that most of time the only informations that you can see on a dask array are its shape and type. You should avoid as much as possible to load the array into memory and define all the operations that you want to perform on an array before launching the actual computation. For example
import xarray as xr dset = xr.open_dataset('dataset.nc', chunks={'time':10}) # This is opened using dask var3 = dset['var'] + dset['var2'] mean = var3.mean(axis=1) mean = mean.compute()
In a normal Python session that does not use the option chunks={'time':10} the operations will be evaluated in every row, and the last one won't make any sense. When using dask, instead, no operation is performed until the command mean.compute() is evaluated.
To start the computation usually you should use the .compute() or .persist() methods. The second one keep the result into memory (handy!) and gives you back the control of the python shell, so it is advisable to use. To load the data into memory you can instead use the .values or .load() attribute and method, respectively.
Using a distributed scheduler in Dask on Mistral
Dask is loaded automatically when using xarray so that maybe some of your computations were already done using it without you noticing. In fact Dask can automatically detect the machine computation capabilities and act accordingly. However, in our case we want to be able to use Dask with a distributed configuration. In simple words Dask will do computations using a scheduler, that is a process that take care of organising the work of many workers which in a cluster architecture are usually equivalent to the nodes. Scheduler and workers can be started on Mistral using different strategies.
The first, easiest one, employs dask-jobqueue.
from dask_jobqueue import SLURMCluster cluster = SLURMCluster(queue='compute2', cores=36, memory='64 GB', project='bm0682', walltime='02:00:00',local_directory='/work/mh0731/m300382/dask/', job_extra=['-o /work/mh0731/m300382/dask/LOG_worker_%j.o', '-e /work/mh0731/m300382/dask/LOG_worker_%j.o'])
What SLURMCluster does is to prepare many scripts which are then submitted automatically to the machine to reserve as many nodes as you want. You can check what the script looks like doing print(cluster.job_script()). To reserve 4 nodes you just have to scale the cluster and connect Dask to it
cluster.scale(4) from dask.distributed import Client client = Client(cluster)
What is amazing in this is that the distributed part of Dask can balance the load depending on how many nodes you have available, which you can easily change even afterwards by scaling again the cluster cluster.scale(8). The drawback is that one job has to be submitted per node, which may be problematic depending on the resources of your group.
Remember to launch the jupyter notebook using the -A option so that you reserved a node on the machine otherwise the notebook won't be able to connect to the workers as mistralpp is not in the same network.
For this reason the jobs can be launched manually using dask-mpi, which is the second method displayed here. In order to do that you can use the following bash script:
just make sure to create a folder, in my case /work/mh0731/m300382/dask, where all the log and temporary files are store, and then launch the script as you would do sbatch -N number_nodes launch_dask_distributed.sh. dask-mpi will use MPI library just to communicate with the nodes and start many workers and a scheduler which are then connected together. Once everything is running smoothly you should connect to the scheduler using this simple snippet in your notebook
import json data=[] with open('/work/mh0731/m300382/dask/scheduler.json') as f: data = json.load(f) scheduler_address=data['address'] from dask.distributed import Client client = Client(scheduler_address)
Using this method only one job will be launched on the machine and you'll be more flexible in choosing a larger number of nodes.
Regardless of which method you use in the end you'll have a client object which Dask will use to distribute the computation. Once the client is started you can use the dashboard to check the status of your cluster and of the computation that you'll eventually launch. This can be usually reached on localhost:8787 (8787 is the default port but it may vary) on your browser but since the scheduler lies on Mistral we have to make a ssh tunnel to make sure that this is locally accessible. Luckily this is quite easy:
ssh -N -L 8787:$1:8787 m300382@mistralpp.dkrz.de
where $1 is the address of your scheduler which will be printed if you just do client. It should be something like 10.50.40.17. If the address is ok when you type in your browser http://localhost:8787 you should get something like this.
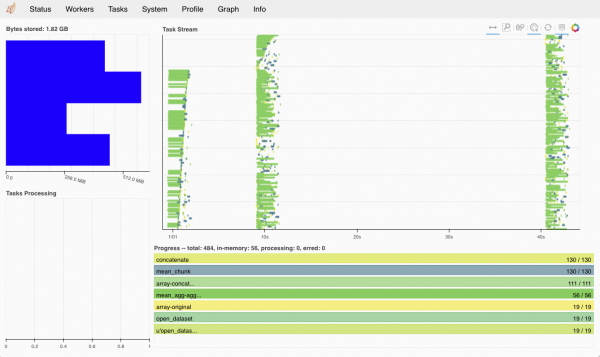 More info on how to use the dashboard can be found here http://docs.dask.org/en/latest/diagnostics-distributed.html.
More info on how to use the dashboard can be found here http://docs.dask.org/en/latest/diagnostics-distributed.html.
Usage example
The example can be found on Mistral at /work/mh0731/m300382/dask/dask_example.ipynb (although it will probably be moved in the future).
Now that we have everything sorted out we can use the distributed computation to compute the average of rain rate over the entire domain of Germany for one of the days simulated (https://code.mpimet.mpg.de/projects/icon-lem/wiki/HDCP2_DE). We start by listing all the files that we need
import numpy as np from glob import glob import matplotlib.pyplot as plt import xarray as xr import dask.array as da import pandas as pd main_folder='/work/bm0834/k203095/ICON_LEM_DE/' file_prefix='2d_cloud_day' domain='DOM01' folders=['20160529-default'] variable='rain_gsp_rate' filenames=[] for folder in folders: filenames=filenames+(sorted(glob(main_folder+folder+'/DATA/'+file_prefix+'_'+domain+'*')))
And then read the single file and concatenate all together
temps=[xr.open_dataset(fn)[variable] for fn in filenames] arrays=[da.from_array(t, chunks=(chunk_time, chunk_space)) for t in temps] var=da.concatenate(arrays, axis=0)
Note again that until now we didn't perform any computation but just told Dask where the data should be found and what are the dimensions of the chunks. Just take one moment to appreciate how flexible and lightweight this is. Data may be scattered in different folders over the machine and without copying them we're just able to merge them and do computation, which will be parallelized automatically depending on how the data is organized.
We can start the computation by doing
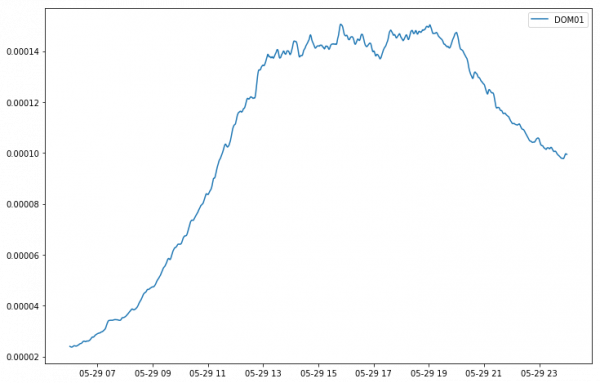
mean = var.mean(axis=1).persist()
If you look into the dashboard you'll see all the chunks being processed, something like this.
Once the computation is done you can save the result by loading into memory
mean = mean.compute() array([2.4039342e-05, 2.3902332e-05, 2.3811526e-05, ..., 9.9697572e-05, 9.9602083e-05, 9.9465833e-05], dtype=float32)
which we can then easily plot.
plt.figure(figsize=(12, 8)) time=pd.date_range(start='2016-05-29 06:00:00', end='2016-05-30 00:00:00', periods=var.shape[0]) plt.plot(time, mean, label='DOM01') plt.legend() plt.show()
Instead of using dask.array we can merge the datasets using xarray directly but specifying the chunks size when loading the dataset. This should work most of the time and it is more flexible but fails if the input files are too large.
temps=[xr.open_dataset(fn, chunks={'time':50}) for fn in filenames] dset = xr.concat(temps, dim='time') dset[variable].mean(axis=1).compute()
Caveats/problems/suggestions
The first and most important:
Dask is not always the answer. If your data fits into memory loading and processing the data with Dask will make everything slower. This is because when the dataset is not big the advantage of processing every chunk separately is overcomed by the intrinsic limited speed of transmitting the results of the computation between the workers. This is nothing new and the same problem that is encountered in parallel computation, the so-called scaling. So, as always, think before computing and processing!
xr.open_mfdataset can be used in place of xr.open_dataset to open multiple files BUT in this case every file will be loaded as single chunk with Dask. Thus the merged array will not be processed as one and the memory could get filled if the files are large. We do not advise to use this method.
Progress of a computation can be tracked also without the dashboard using the progress class of dask.distributed
Dask distributed can be run also on your laptop using the class LocalCluster.
GRIB files can be opened with xarray when cfgrib (https://github.com/ecmwf/cfgrib) is installed. Unfortunately GRIB files still remain GRIB files so they're not really flexible. So in theory you can use the method shown before also with grib files, but it will not always work. More importantly DYAMOND files are written in a compressed GRIB2 format. Luckily the eccodes library shipped with conda-forge comes with the de-compression library installed but it will still hinder the performance of reading the files. So you may want to convert them to netcdf before analysing it. I know, it sounds stupid but it works.
For a xarray-native workaround to the above issue with grib1 output and CDO stream.codes files (found in exp_id/log), open grb files with xr.open_mfdataset(engine=“cfgrib”), and rename variables based on pd.read_fwf(stream.codes, sep=“\t”, header=None, names=[“param”,“nlev”,“varname”,“some1”,“some2”,“long_name_unit”], index_col=0) looping over variables v and renaming based on ds[v].attrs['GRIB_paramId'] as demonstrated in https://gist.github.com/aaronspring/bf5139a718bbd607dcb95a5222a9d0e8.