ICON quick start guide
So you want to work with ICON?
Although documentation regarding the ICON model has been steadily increasing in the latest years new PhD or master students still struggle to set up the first simulation. This quick guide will help you in getting started from getting the code to start your first simulation. It is not meant to be a comprehensive guide but rather a collection of hacks/tricks which hopefully will get you started. Attention will be paid on the ICON-LEM branch of the model, which allows one to run realistic or idealized simulations at grid spacing typically smaller than 5 km. Thus, this tutorial is NOT intended to be used for climate simulations or ICON-NWP simulations in general, for which tutorials are linked in How to?.
What you need
Usually running the model is the easiest step. Depending on your configuration you may need to pre-process input data, which is instead the most annoying part. For the idealized simulations performed over the Torus domain (periodic boundary conditions, see https://agupubs.onlinelibrary.wiley.com/doi/10.1002/2015MS000431) the input data is minimal. To briefly summarise
In terms of code a maximum of 3 different repositories is needed, that is
-
-
-
The EXTPAR repository is needed to create external parameters like soil type, orography and remap them to the input grid. This is useful for both realistic simulations and idealized simulations which use the land-surface parametrization instead than constant SSTs or fixed surface fluxes. This repository is, at the time of writing, available as module on Mistral but the old version can be directly copied from many folders on Mistral (e.g. /work/bm0834/k203095/pool/EXTPAR/).
Getting and compiling the code
Please refer to the wikis of the relative projects on the redmine for more information about obtaining and compiling the code.
ICON
There are many different versions of ICON: a NWP branch used at DWD, a deprecated ICON-LEM version and the main ICON-AES repository used in the atmosphere department. Make sure to load the latest version of git on your machine
module load git/2.9.0
and then clone the repository and checkout the branch that you want to work on (we'll just use the master).
git clone --recursive git@git.mpimet.mpg.de:icon-aes.git
cd icon-aes
git checkout -b icon-aes-master remotes/origin/master
git submodule update
This will create a new local branch icon-aes-master which is tracking the remote master located on the repository. This way you can always get new updates by doing git fetch and git pull. Have a look here for more information on git (https://code.mpimet.mpg.de/projects/icon/wiki/Notes_on_Git). Now you can go ahead and compile
./configure --with-fortran=intel --disable-ocean --disable-jsbach
./build_command
This will compile the code using the ifort compiler and disabling the ocean and jsbach physics (we don't need those for our limited area simulations). Take a look at all the configure options if you need to change something.
A compiled version of ICON tools is on levante and you should try to obtain it from there. If you download ICON tools using git, then once you have downloaded it using git go into the icontools folder. On Mistral the following configuration works, but always refer to the guide since the configuration to compile the icontools continuously changes.
First, go into the Makefile and change the configuration to use eccodes instead than grib-api:
GFORTRAN_OPENMPI_GRIBAPIROOT = /sw/rhel6-x64/eccodes/eccodes-2.5.0-gcc48/
GFORTRAN_OPENMPI_LIBS = \
-L../lib ${TOOL_LIBS} ${ADDITIONAL_LIBS} -L${GFORTRAN_OPENMPI_NETCDFFROOT}/lib \
-L${GFORTRAN_OPENMPI_NETCDFCROOT}/lib -L${GFORTRAN_OPENMPI_GRIBAPIROOT}/lib \
-L${GFORTRAN_OPENMPI_HDF5ROOT}/lib \
-lgfortran -lnetcdff -lnetcdf -lhdf5_hl -lhdf5 -lm -lz -leccodes -ljasper
Remember that you'll need to add these lines in the runscript afterwards as written in the Makefile (see create initial condition and boundary conditions section):
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/sw/rhel6-x64/netcdf/netcdf_fortran-4.4.3-gcc62/lib/
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/sw/rhel6-x64/netcdf/netcdf_c-4.4.1.1-gcc48/lib
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/sw/rhel6-x64/hdf5/hdf5-1.8.14-gcc48/lib
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/sw/rhel6-x64/eccodes/eccodes-2.5.0-gcc48/lib/
Finally compile
module load gcc/7.1.0
module load netcdf_c/4.3.2-gcc48
make gfortran_openmpi_generic
If everything goes well you should see new executables create in the folder: icondelaunay_mpi, icongpi, icongridgen, iconremap_mpi, iconsub_mpi.
If you manage to find a icontools version already compiled on Mistral it was probably made using ifort and it will work without these hacks. Unfortunately the target to compile icontools with ifort has been removed.
ICON Grid Generator
git clone --recursive git@git.mpimet.mpg.de:GridGenerator.git
cd GridGenerator
./configure --with-fortran=intel --with-openmp --with-flags=hiopt
./build_command
Doing this you should be able to run the grid generator scripts either interactively or to submit them to the queue.
Now that the model is ready we should take a look on how to create the input data. The amount of data specifically depends on the setup that you want to run. I will assume that you already compiled the relevant repositories like the ICON tools and the ICON grid generator.
Create the grid
The ICON Grid generator wiki (https://code.mpimet.mpg.de/projects/icon-grid-generator/wiki) contains a lot of examples. To generate the grid you just have to copy one of the script from the GridGenerator/run/grid_creators folder to the GridGenerator/run/ folder and then make a run version using GridGenerator/make_runscripts. This will take care of defining the correct settings according to the ones that were used at compilation time. Grids are created in the /grids directory and should then copied or moved to the experiment directory where we'll launch the simulation.
Remember to compile the GridGenerator with the options advised on the wiki and to submit as job the grid generation script, especially if your grids have a lot of points. If the grid that you're trying to generate does not contain a lot of points you can just run the script interactively ./grid.generate.run but you're using shared resources on the login node that you're connected to.
Torus domain
This grid is flat, has doubly periodic boundary conditions and is used for idealized LES simulations in ICON-LEM (see https://agupubs.onlinelibrary.wiley.com/doi/10.1002/2015MS000431). The relevant script is grid.create_torus_grid. Just modify the line
create_torus 100 100 500
to contain, respectively, the number of points in the x, y directions and the edge length (grid spacing) in meters. Then run the script.
Limited-area grid
These are grids covering a fraction of the globe. They can be created following to main methodologies:
Cut an area from an available global grid: this is the setup employed in many projects like HD(CP)2. However, you need an input global grid to start with.
Create a limited area grid directly: this create higher quality grids and is the default at the DWD for the NWP setup. However, older setups at MPI did not use this method, which is then not fully supported, yet. Grids created with the online grid generator are also of this type.
In the following, we'll attempt to create a limited grid over Italy which also include some nests. Our target resolution is about 1 km so not only we need to cut a grid from the global domain but also to refine the resolution. In order to use the first method you can modify the script grid.create_limited_area_gridsby activating the option make_germany_hdcp2=“true” and use the relevant part of the script (but you can still add your own option in the script…):
grid.create_limited_area_grids
grid.create_limited_area_grids
set_no_optimization_grids
no_of_conditions=1
patch_shape=$lonlat_rectangle_condition
patch_center_x=12.
patch_center_y=42.
patch_rectangle_xradious=6.5
patch_rectangle_yradious=7.
out_file_prefix="italy_grid"
input_file="Global_Icos_4932m_springOpt.nc"
output_file="${out_file_prefix}<resolution>.nc"
create_hierarchy=".true."
refine_depth=2
optimize_depth=2
create_refined_patches
With patch_shape the shape of the region of interest can be chosen which is either rectangular (lonlat_rectangle_condition) or circular (circle_shape). The center in x-(lon)direction and y-(lat)direction, respectively is set via patch_center_x and patch_center_y, respectively. The unit is degrees lon and degrees lat, respectively. The width and height (or radius in case of circular shape) is set via patch_rectangle_xradious and patch_rectangle_yradious, again in degrees lon and degrees lat. The input global grid (in this case Global_Icos_4932m_springOpt.nc) can be either taken from ones that are already available on Mistral or generated using the relevant script (grid.create_Global_Icosahedron_grids).
refine_depth defines the number of refinements that shall be created, thus increasing the resolution. In the example above, refine_depth=2 creates two refinements with halved resolution: since we are starting from a global domain with approximately 5 km grid spacing we'll get two refinements at 2.5 and 1.25 km grid spacing, respectively, italy_grid2462m.nc and italy_grid1231m.nc, our target resolution. We'll use this grid in the following sections as parent grid to create the nests. The option create_hierarchy=“.true.” enables the generation of refinements of the grid.
Note that if you don't need to refine the global grid but just cut (thus keeping resolution constant) everything becomes even simpler and you can even combine the generation of the global grid with the cut of the local grid (if resolution is not too high, otherwise the process should be separated as the generation of global grids at high resolution take a while). For example, to obtain a 40 km grid that spans the Tropical Atlantic you can just do (thanks to Leonidas for the hint):
grid.create_limited_area_grids
grid.create_limited_area_grids
# create global grids
set_spr_optimization_grids
base_file_name="Global_Icos_<resolution>.nc"
max_resolution=40000
create_icosahedron_grids
# create first patch
cut_lonlat_rectangle_grid Global_Icos_0039km.nc tropical_atlantic_grid.nc -14.5 4 85 54
To use the second method described before (cut without using the global grid) activate make_Guido_grid=“true” or make_Aiko_grid=“true” and modify the relevant parameters. This method should be used only if you don't need to nest a limited-area grid into a global.
Nested grids
Nested grids are useful if one wants to run different refinements at the same time nested into each other. A parent grid has to be generated beforehand, using the method shown in the previous section and will be taken as input for the nests. The relevant script is grid.create_icon_nested_grids. There are many options in this script but we will only look into the example make_test_hdcp2_nestpatches=“true” where we'll attempt to create 2 nests with a parent domain. In order to do this, we also have to use the parent grid created before (italy_grid1231m.nc) which will be overwritten by the script, so that we need to copy it beforehand.
grid.create_icon_nested_grids
grid.create_icon_nested_grids
# create nests
rm italy_grid_nested_*.nc
cp italy_grid1231m.nc italy_grid_nested_1231m.nc
cut=$lonlat_rectangle_condition
inner=$inner_cells_condition
inner_cells_depth=13
no_of_patches=3
patchNameList=(italy_grid_nested_1231m.nc italy_grid_nested_616m.nc italy_grid_nested_308m.nc )
patchIDList=( 1 2 3 )
patchParentIDList=( 0 1 2 )
patchShapeList=( none $inner $cut )
patchCenterXList=( none none 10.5 )
patchCenterYList=( none none 43. )
rectangleXradiousList=(none none 1.5 )
rectangleYradiousList=(none none 1.5 )
boundary_indexing_depth=12
set_hdcp2_optimization_grids
create_hierarchical_grids
In this example the first nest, italy_grid_nested_616m.nc, will be created just inside the parent grid, italy_grid_nested_1231m.nc, so that it will cover roughly the same area. For the innermost domain italy_grid_nested_308m.nc, instead, we define a lat-lon box (centered at 10.5, 43 and spanning 1.5 degrees in both directions), also because covering the whole domain with a 300 m grid would be quite expensive. Of course you can change the setup to have less domains and different conditions.
The parameters boundary_indexing_depth and inner_cells_depth refer to the relationship between parent and child grid and are explained by means of the image below.
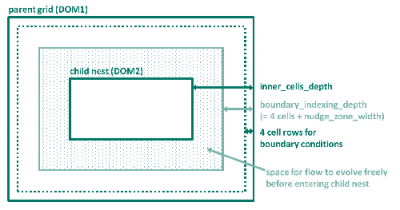
inner_cells_depth is the total number of cell rows between the parent grid and its child nest (and between subsequent child nests, respectively). The parameter boundary_indexing_depth denotes the number of cell rows which the grid generator reserves for boundary conditions and nudging. The number of cell rows for boundary conditions is hard coded in ICON to the value of 4. The number of cell rows for nudging is specified via the ICON namelist parameter nudge_zone_width to be defined in the namelist interpol_nml. This means that inner_cells_depth is the sum of the number of cell rows for boundary conditions (4), nudge_zone_width and number of cell rows where the flow can freely evolve in the parent grid before entering the nest.
Plot the grids
It is a good idea to always check the grid extents before proceeding further. Here is a small Python script that prints the extents of the grid and draws a map with the grid overlaid. If the correct backend is loaded you can zoom into the map and check every boundary in detail. Just run the script with the grid name, e.g. python plot_grid.py grid_file_name.nc
- plot_grid.py
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from mpl_toolkits.basemap import Basemap # Import the Basemap toolkit
import xarray as xr
import sys
file = sys.argv[1:]
dset = xr.open_dataset(str(file[0]))
lon_min = np.asscalar(np.min(dset.clon))
lon_max = np.asscalar(np.max(dset.clon))
lat_min = np.asscalar(np.min(dset.clat))
lat_max = np.asscalar(np.max(dset.clat))
print('Longitude min. %7.4f, max. %7.4f' % (np.rad2deg(lon_min),np.rad2deg(lon_max)) )
print('Latitude min. %7.4f, max. %7.4f' % (np.rad2deg(lat_min),np.rad2deg(lat_max)) )
def get_projection(dset):
lon_center = np.rad2deg(dset.clon.values).mean()
lat_center = np.rad2deg(dset.clat.values).mean()
x, y = np.rad2deg(dset.clon.values), np.rad2deg(dset.clat.values)
m = Basemap(projection='nsper',lon_0=lon_center, lat_0=lat_center,
resolution='l',satellite_height=15000000)
m.drawcountries(linewidth=0.5, color='black')
m.shadedrelief()
x, y = m(x, y)
return(m, x, y)
fig = plt.figure(figsize=(10,10))
m, x, y = get_projection(dset)
z = np.ones(x.shape)
m.contourf(x, y, z, tri=True, alpha=0.4, levels=(1.,2.), colors='red')
plt.show(block=True)
Create the extpar file
At the time of writing the extpar package is undergoing big changes and a new version has been developed at MPI and will be available on Mistral as module in the future. Please refer to Luis Kornblueh in the future.
For realistic simulations, geographically localized data sets like the topographic height of the earth surface, the plant cover or the distribution of land and sea need to be interpolated to the grid used in the simulation beforehand. The EXTPAR software system (EXTPAR - External Parameter for Numerical Weather Prediction and Climate Application) developed at DWD (Jürgen Helmert) and Meteo Swiss is used. There is no publicly available repository for this software system. It can be obtained on request in case external data shall be produced for a certain setup. (A copy of Version V2_4 of EXTPAR is located on Mistral in pool of the HD(CP)2 project: /work/bm0834/k203095/pool/EXTPAR/.) As input it requires the grids for all domains and external data sets and it creates an extpar file for every domain.
The input data are
| Variable | Dataset | Source | Resolution | Notes |
| Global topography | GLOBE orography | NOAA/NGDC | 30” |
| Topography | ASTER orography | METI/NASA | 1” | limited domain: 60°N - 60°S |
| Surface cover type | Globcover 2009 | ESA | 10” |
| Land classes | GLC2000 land use | JRC Ispra | 30” |
| Land classes | GLCC land use | USGS | 30” |
| Soil type | DSMW Digital Soil Map of the World | FAO | 5’ |
| Soil type | HWSD Harmonized World Soil Database | FAO/IIASA/ISRIC/ISSCAS/JRC | 30” |
| Normalized Difference Vegetation Index | NDVI Climatotology, SEAWiFS | NASA/GSFC | 2.5’ |
| Temperature climatology for deep soil layers | CRU near surface climatology | CRU University of East Anglia | 0.5 degree |
| Aerosol Optical thickness | Aerosol Optical thickness | NASA/GISS | 4×5 degree |
| Aerosol Optical thickness | AeroCom Global AOD data | AeroCom Project | 1 degree |
| Lake cover | Global lake database (GLDB) | DWD/RSHU/MeteoFrance | 30” |
| Albedo | MODIS albedo | NASA | 5’ |
The ASTER topography saved on Mistral only has tiles covering Western Europe. If you need to create a high-resolution extpar file for an area located outside you may need to download the data beforehand or consider using the low-resolution GLOBE topography dataset.
The code is already compiled and can be run directly without issues. An example script is attached.
Bash script to generate an extpar file
Bash script to generate an extpar file
- run_extpar_icon_mistral.sh
#!/bin/bash
#
# Generate external parameters for HDCP2 simulations
# using GLOBCOVER2009, FAO DSMW, ASTER, Lake Database
# 06.2014 Daniel Klocke
# Adapted for Blizzard by Anurag Dipankar
# Adapted for mistral by Pavan Siligam
#=====================================
# slrum batch job parameters
# these are specified in cylc suite.rc
#-------------------------------------
#SBATCH --account=bm0682
#SBATCH --job-name=extpar_icon_mistral.run
#SBATCH --partition=compute2
#SBATCH --nodes=1
#SBATCH --threads-per-core=2
#SBATCH --output=LOG.extpar_icon_mistral.run.%j.o
#SBATCH --error=LOG.extpar_icon_mistral.run.%j.o
#SBATCH --exclusive
# #
#========================================
# GENERAL REMARKS (Guido)
# Depending on the selection of globcover and orography things can get weird.
# so please not the following:
# -ASTER topography only goes to 35N, if you want to go south you won't get far
# -GLOBCOVER HDCP2 only works for northern Europe
#
# REMOVE EVERYTHING FROM EXP FOLDER BEFORE LAUNCHING WHEN CHANGING GRID!!!!
#-----------------------------------------------------------------------------
# openmp environment variables
# ----------------------------
export OMP_NUM_THREADS=24
export OMP_SCHEDULE=static,16
export OMP_DYNAMIC="false"
export OMP_STACKSIZE=128M
ulimit -s unlimited
set -e
SWROOT=/sw/rhel6-x64
GRIBROOT=${SWROOT}/grib_api/grib_api-1.13.0-intel14
export GRIB_DEFINITION_PATH=${GRIBROOT}/share/grib_api/definitions/:${GRIBROOT}/share/grib_api/definitions/grib2/localConcepts/edzw
export GRIB_SAMPLE_PATH=${GRIBROOT}/share/grib_api/samples
data_dir=/work/bm0834/k203095/pool/EXTPAR_INPUT
#For input grid
filename=hdcp2_R2B07-grid
#
#Where Binaries are
progdir=/work/mh0731/m300382/extpar_lem/bin
#
workdir=/work/mh0731/m300382/extpar_lem/exp
#
#Grid dir
#icon_grid_dir=/work/mh0731/m300382/icon-GridGenerator-dev/grids
icon_grid_dir=/scratch/m/m300382/temp
#
# set target grid definition
icon_grid_file=${filename}.nc
if [[ ! -d ${workdir} ]] ; then
mkdir -p ${workdir}
fi
cd ${workdir}
pwd
binary_extpar_consistency_check=extpar_consistency_check.exe
binary_aot=extpar_aot_to_buffer.exe
binary_tclim=extpar_cru_to_buffer.exe
binary_lu=extpar_landuse_to_buffer.exe
binary_topo=extpar_topo_to_buffer.exe
binary_ndvi=extpar_ndvi_to_buffer.exe
binary_soil=extpar_soil_to_buffer.exe
binary_flake=extpar_flake_to_buffer.exe
binary_alb=extpar_alb_to_buffer.exe
cat > INPUT_grid_org << EOF_go
&GRID_DEF
igrid_type = 1,
domain_def_namelist='INPUT_ICON_GRID'
/
EOF_go
cat > INPUT_ICON_GRID << EOF5
&icon_grid_info
icon_grid_dir='${icon_grid_dir}',
icon_grid_nc_file ='${filename}.nc'
/
EOF5
#---
grib_output_filename="extpar_${filename}.g2"
netcdf_output_filename="extpar_${filename}.nc"
grib_sample='GRIB2'
echo $netcdf_output_filename
echo $grib_output_filename
#----
raw_data_alb='month_alb.nc'
raw_data_alnid='month_alnid.nc'
raw_data_aluvd='month_aluvd.nc'
buffer_alb='month_alb_BUFFER.nc'
output_alb='month_alb_extpar_ICON.nc'
raw_data_aot='aerosol_optical_thickness.nc'
buffer_aot='extpar_aot_BUFFER.nc'
output_aot='aot_extpar_ICON.nc'
raw_data_tclim_coarse='absolute_hadcrut3.nc'
raw_data_tclim_fine='CRU_T2M_SURF_clim.nc'
buffer_tclim='crutemp_climF_extpar_BUFFER.nc'
output_tclim='crutemp_climF_extpar_ICON.nc'
raw_data_glc2000='glc2000_byte.nc'
buffer_glc2000='extpar_landuse_BUFFER.nc'
output_glc2000='extpar_landuse_ICON.nc'
raw_data_glcc='glcc_usgs_class_byte.nc'
buffer_glcc='glcc_landuse_BUFFER.nc'
output_glcc='glcc_landuse_ICON.nc'
raw_data_globcover_0='GLOBCOVER_0_16bit.nc'
raw_data_globcover_1='GLOBCOVER_1_16bit.nc'
raw_data_globcover_2='GLOBCOVER_2_16bit.nc'
raw_data_globcover_3='GLOBCOVER_3_16bit.nc'
raw_data_globcover_4='GLOBCOVER_4_16bit.nc'
raw_data_globcover_5='GLOBCOVER_5_16bit.nc'
raw_data_globcover='GLOBCOVER_USERSPECIF_HDCP2.nc'
buffer_lu='extpar_landuse_BUFFER.nc'
output_lu='extpar_landuse_ICON.nc'
raw_data_globe_A10='GLOBE_A10.nc'
raw_data_globe_B10='GLOBE_B10.nc'
raw_data_globe_C10='GLOBE_C10.nc'
raw_data_globe_D10='GLOBE_D10.nc'
raw_data_globe_E10='GLOBE_E10.nc'
raw_data_globe_F10='GLOBE_F10.nc'
raw_data_globe_G10='GLOBE_G10.nc'
raw_data_globe_H10='GLOBE_H10.nc'
raw_data_globe_I10='GLOBE_I10.nc'
raw_data_globe_J10='GLOBE_J10.nc'
raw_data_globe_K10='GLOBE_K10.nc'
raw_data_globe_L10='GLOBE_L10.nc'
raw_data_globe_M10='GLOBE_M10.nc'
raw_data_globe_N10='GLOBE_N10.nc'
raw_data_globe_O10='GLOBE_O10.nc'
raw_data_globe_P10='GLOBE_P10.nc'
buffer_topo='topography_BUFFER.nc'
output_topo='topography_ICON.nc'
raw_data_ndvi='NDVI_1998_2003.nc'
buffer_ndvi='NDVI_BUFFER.nc'
output_ndvi='ndvi_extpar_ICON.nc'
raw_data_soil_FAO='FAO_DSMW_DP.nc'
raw_data_soil_HWSD='HWSD0_30_texture_2.nc'
raw_data_deep_soil='HWSD30_100_texture_2.nc'
buffer_soil='SOIL_BUFFER.nc'
output_soil='SOIL_ICON.nc'
raw_lookup_table_HWSD='LU_TAB_HWSD_UF.data'
raw_HWSD_data='HWSD_DATA_COSMO.data'
raw_HWSD_data_deep='HWSD_DATA_COSMO_S.data'
raw_HWSD_data_extpar='HWSD_DATA_COSMO_EXTPAR.asc'
raw_data_flake='lakedepth.nc'
buffer_flake='flake_BUFFER.nc'
output_flake='ext_par_flake_ICON.nc'
# create input namelists
cat > INPUT_AOT << EOF_aot
&aerosol_raw_data
raw_data_aot_path='',
raw_data_aot_filename='${raw_data_aot}'
/
&aerosol_io_extpar
aot_buffer_file='${buffer_aot}',
aot_output_file='${output_aot}'
/
EOF_aot
#---
cat > INPUT_TCLIM << EOF_tclim
&t_clim_raw_data
raw_data_t_clim_path='',
raw_data_t_clim_filename='${raw_data_tclim_coarse}'
raw_data_t_id = 2
/
&t_clim_io_extpar
t_clim_buffer_file='crutemp_climC_extpar_BUFFER.nc',
t_clim_output_file='crutemp_climC_extpar_ICON.nc'
/
EOF_tclim
#---
# &lu_raw_data
# raw_data_lu_path='',
# raw_data_lu_filename='${raw_data_globcover}',
# i_landuse_data=1,
# ilookup_table_lu=1,
# ntiles_globcover=1
# /
cat > INPUT_LU << EOF_lu
&lu_raw_data
raw_data_lu_path='',
raw_data_lu_filename='${raw_data_globcover_0}' '${raw_data_globcover_1}' '${raw_data_globcover_2}' '${raw_data_globcover_3}' '${raw_data_globcover_4}' '${raw_data_globcover_5}',
i_landuse_data=1,
ilookup_table_lu=1,
ntiles_globcover=6
/
&lu_io_extpar
lu_buffer_file='${buffer_lu}',
lu_output_file='${output_lu}'
/
&glcc_raw_data
raw_data_glcc_path='',
raw_data_glcc_filename='${raw_data_glcc}'
/
&glcc_io_extpar
glcc_buffer_file='${buffer_glcc}',
glcc_output_file='${output_glcc}'
/
EOF_lu
#---
#cat > INPUT_ORO << EOF_oro
#&orography_io_extpar
#orography_buffer_file='${buffer_topo}',
#orography_output_file='${output_topo}'
#/
#&orography_raw_data
#itopo_type = 2
#lsso_param = .TRUE.,
#raw_data_orography_path='',
#ntiles_column = 2,
#ntiles_row = 4,
#topo_files = 'topo.ASTER_orig_T006.nc' 'topo.ASTER_orig_T007.nc' 'topo.ASTER_orig_T018.nc' 'topo.ASTER_orig_T019.nc' 'topo.ASTER_orig_T030.nc' 'topo.ASTER_orig_T031.nc ' 'topo.ASTER_orig_T042.nc' 'topo.ASTER_orig_T043.nc'
#/
#EOF_oro
cat > INPUT_ORO << EOF_oro
&orography_io_extpar
orography_buffer_file='${buffer_topo}',
orography_output_file='${output_topo}'
/
&orography_raw_data
itopo_type = 1
lsso_param = .TRUE.,
raw_data_orography_path='',
ntiles_column = 4,
ntiles_row = 4,
topo_files = ${raw_data_globe_A10} ${raw_data_globe_B10} ${raw_data_globe_C10} ${raw_data_globe_D10} ${raw_data_globe_E10} ${raw_data_globe_F10} ${raw_data_globe_G10} ${raw_data_globe_H10} ${raw_data_globe_I10} ${raw_data_globe_J10} ${raw_data_globe_K10} ${raw_data_globe_L10} ${raw_data_globe_M10} ${raw_data_globe_N10} ${raw_data_globe_O10} ${raw_data_globe_P10}
/
EOF_oro
#----
cat > INPUT_OROSMOOTH << EOF_orosmooth
&orography_smoothing
lfilter_oro=.FALSE.
/
EOF_orosmooth
#---
cat > INPUT_RADTOPO << EOF_rad
&radtopo
lradtopo=.FALSE.,
nhori=24,
/
EOF_rad
#---
#---
cat > INPUT_NDVI << EOF_ndvi
&ndvi_raw_data
raw_data_ndvi_path='',
raw_data_ndvi_filename='${raw_data_ndvi}'
/
&ndvi_io_extpar
ndvi_buffer_file='${buffer_ndvi}',
ndvi_output_file='${output_ndvi}'
/
EOF_ndvi
#---
cat > INPUT_SOIL << EOF_soil
&soil_raw_data
isoil_data = 2,
ldeep_soil = .false.,
raw_data_soil_path='',
raw_data_soil_filename='${raw_data_soil_HWSD}'
raw_data_deep_soil_filename='${raw_data_deep_soil}'
/
&soil_io_extpar
soil_buffer_file='${buffer_soil}',
soil_output_file='${output_soil}'
/
&HWSD_index_files
path_HWSD_index_files='',
lookup_table_HWSD='${raw_lookup_table_HWSD}',
HWSD_data='${raw_HWSD_data}',
HWSD_data_deep='${raw_HWSD_data_deep}',
HWSD_data_extpar='${raw_HWSD_data_extpar}'
/
EOF_soil
#---
cat > INPUT_FLAKE << EOF_flake
&flake_raw_data
raw_data_flake_path='',
raw_data_flake_filename='${raw_data_flake}'
/
&flake_io_extpar
flake_buffer_file='${buffer_flake}'
flake_output_file='${output_flake}'
/
EOF_flake
#---
cat > INPUT_ALB << EOF_alb
&alb_raw_data
raw_data_alb_path='',
raw_data_alb_filename='${raw_data_alb}'
/
&alnid_raw_data
raw_data_alb_path='',
raw_data_alnid_filename='${raw_data_alnid}'
/
&aluvd_raw_data
raw_data_alb_path='',
raw_data_aluvd_filename='${raw_data_aluvd}'
/
&alb_io_extpar
alb_buffer_file='${buffer_alb}',
alb_output_file='${output_alb}'
/
&alb_source_file
alb_source='al'
alnid_source='alnid'
aluvd_source='aluvd'
/
EOF_alb
# consistency check
cat > INPUT_CHECK << EOF_check
&extpar_consistency_check_io
grib_output_filename='${grib_output_filename}',
netcdf_output_filename='${netcdf_output_filename}',
orography_buffer_file='${buffer_topo}',
soil_buffer_file='${buffer_soil}',
lu_buffer_file='${buffer_lu}',
glcc_buffer_file='${buffer_glcc}',
flake_buffer_file='${buffer_flake}',
ndvi_buffer_file='${buffer_ndvi}',
t_clim_buffer_file='${buffer_tclim}',
aot_buffer_file='${buffer_aot}',
alb_buffer_file='${buffer_alb}',
i_lsm_data=1,
land_sea_mask_file="",
number_special_points=0
/
EOF_check
# link raw data files to local workdir
ln -s -f ${data_dir}/${raw_data_alb}
ln -s -f ${data_dir}/${raw_data_alnid}
ln -s -f ${data_dir}/${raw_data_aluvd}
ln -s -f ${data_dir}/${raw_data_aot}
ln -s -f ${data_dir}/${raw_data_tclim_coarse}
ln -s -f ${data_dir}/${raw_data_tclim_fine}
ln -s -f ${data_dir}/${raw_data_glc2000}
ln -s -f ${data_dir}/${raw_data_glcc}
ln -s -f ${data_dir}/${raw_data_globcover_0}
ln -s -f ${data_dir}/${raw_data_globcover_1}
ln -s -f ${data_dir}/${raw_data_globcover_2}
ln -s -f ${data_dir}/${raw_data_globcover_3}
ln -s -f ${data_dir}/${raw_data_globcover_4}
ln -s -f ${data_dir}/${raw_data_globcover_5}
ln -s -f ${data_dir}/${raw_data_globcover}
ln -s -f ${data_dir}/${raw_data_globe_A10}
ln -s -f ${data_dir}/${raw_data_globe_B10}
ln -s -f ${data_dir}/${raw_data_globe_C10}
ln -s -f ${data_dir}/${raw_data_globe_D10}
ln -s -f ${data_dir}/${raw_data_globe_E10}
ln -s -f ${data_dir}/${raw_data_globe_F10}
ln -s -f ${data_dir}/${raw_data_globe_G10}
ln -s -f ${data_dir}/${raw_data_globe_H10}
ln -s -f ${data_dir}/${raw_data_globe_I10}
ln -s -f ${data_dir}/${raw_data_globe_J10}
ln -s -f ${data_dir}/${raw_data_globe_K10}
ln -s -f ${data_dir}/${raw_data_globe_L10}
ln -s -f ${data_dir}/${raw_data_globe_M10}
ln -s -f ${data_dir}/${raw_data_globe_N10}
ln -s -f ${data_dir}/${raw_data_globe_O10}
ln -s -f ${data_dir}/${raw_data_globe_P10}
ln -s -f ${data_dir}/topo.ASTER_orig_T006.nc
ln -s -f ${data_dir}/topo.ASTER_orig_T007.nc
ln -s -f ${data_dir}/topo.ASTER_orig_T008.nc
ln -s -f ${data_dir}/topo.ASTER_orig_T018.nc
ln -s -f ${data_dir}/topo.ASTER_orig_T019.nc
ln -s -f ${data_dir}/topo.ASTER_orig_T020.nc
ln -s -f ${data_dir}/topo.ASTER_orig_T030.nc
ln -s -f ${data_dir}/topo.ASTER_orig_T031.nc
ln -s -f ${data_dir}/topo.ASTER_orig_T032.nc
ln -s -f ${data_dir}/topo.ASTER_orig_T042.nc
ln -s -f ${data_dir}/topo.ASTER_orig_T043.nc
ln -s -f ${data_dir}/topo.ASTER_orig_T044.nc
#ln -s -f ${data_dir}/topo.ASTER_orig_T054.nc
#ln -s -f ${data_dir}/topo.ASTER_orig_T055.nc
ln -s -f ${data_dir}/${raw_data_ndvi}
ln -s -f ${data_dir}/${raw_data_soil_FAO}
ln -s -f ${data_dir}/${raw_data_soil_HWSD}
ln -s -f ${data_dir}/${raw_data_deep_soil}
ln -s -f ${data_dir}/${raw_lookup_table_HWSD}
ln -s -f ${data_dir}/${raw_HWSD_data}
ln -s -f ${data_dir}/${raw_HWSD_data_deep}
ln -s -f ${data_dir}/${raw_data_flake}
ln -s -f ${icon_grid_dir}/${icon_grid_file}
# run the programs
# the next seven programs can run independent of each other
time ${progdir}/${binary_tclim}
#---
cat > INPUT_TCLIM << EOF_tclim
&t_clim_raw_data
raw_data_t_clim_path='',
raw_data_t_clim_filename='${raw_data_tclim_fine}',
raw_data_t_id = 1
/
&t_clim_io_extpar
t_clim_buffer_file='${buffer_tclim}',
t_clim_output_file='${output_tclim}'
/
EOF_tclim
time ${progdir}/${binary_lu}
time ${progdir}/${binary_tclim}
time ${progdir}/${binary_alb}
time ${progdir}/${binary_aot}
time ${progdir}/${binary_topo}
time ${progdir}/${binary_ndvi}
time ${progdir}/${binary_soil}
time ${progdir}/${binary_flake}
printenv | grep GRIB_
cat > INPUT_TCLIM_FINAL << EOF_tclim
&t_clim_raw_data
raw_data_t_clim_path='',
raw_data_t_clim_filename='${raw_data_tclim_fine}',
raw_data_t_id = 1
/
&t_clim_io_extpar
t_clim_buffer_file='crutemp_climF_extpar_BUFFER.nc',
t_clim_output_file='crutemp_climC_extpar_BUFFER.nc'
/
EOF_tclim
time ${progdir}/${binary_extpar_consistency_check}
echo 'External parameters for refinement grid of ICON model generated in'
echo $workdir
Note that this script can be run also interactively if the grid is small, i.e. ./run_extpar_icon_mistral_new.sh. In the file only the name of the input grid needs to be changed
#For input grid
filename=hdcp2_R2B07-grid
#Grid dir
icon_grid_dir=/grid_dir/
If necessary also the topography data and the globcover data can be changed. This usually corresponds just to comment/uncomment some parts of the code, for example for the topography the following lines need to be modified
cat > INPUT_ORO << EOF_oro
&orography_io_extpar
orography_buffer_file='${buffer_topo}',
orography_output_file='${output_topo}'
/
&orography_raw_data
itopo_type = 2
lsso_param = .TRUE.,
raw_data_orography_path='',
ntiles_column = 2,
ntiles_row = 5,
topo_files = 'topo.ASTER_orig_T006.nc' 'topo.ASTER_orig_T007.nc' ...........
/
EOF_oro
# cat > INPUT_ORO << EOF_oro
# &orography_io_extpar
# orography_buffer_file='${buffer_topo}',
# orography_output_file='${output_topo}'
# /
# &orography_raw_data
# itopo_type = 1
# lsso_param = .TRUE.,
# raw_data_orography_path='',
# ntiles_column = 4,
# ntiles_row = 4,
# topo_files = ${raw_data_globe_A10} .........
# /
# EOF_oro
When running the script some messages will be printed on screen, no worries! If everything is going well at the end a netcdf file with the same name of the input grid will be generated in the /exp/ directory.
Please make sure to remove everything in the /exp/ directory before launching the script. In case you change grid, the script could fail just because old files are not being replaced in this directory and the error will not be traceable.
As you need a grid file for every domain/nest you'll also need an extpar file for every domain/nest, so remember to generate them and check them before proceeding further.
Take a moment to look at the extpar files before running the simulation. Here is a script which plots the topography and can be run with python plot_extpar.py grid_file.nc.
- plot_extpar.py
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from mpl_toolkits.basemap import Basemap # Import the Basemap toolkit
import xarray as xr
import sys
import matplotlib.colors as colors
file = sys.argv[1:]
dset = xr.open_dataset(str(file[0]))
lon_min = np.asscalar(np.min(dset['lon']))
lon_max = np.asscalar(np.max(dset['lon']))
lat_min = np.asscalar(np.min(dset['lat']))
lat_max = np.asscalar(np.max(dset['lat']))
print('Longitude min. %7.4f, max. %7.4f' % (lon_min, lon_max) )
print('Latitude min. %7.4f, max. %7.4f' % (lat_min,lat_max) )
def get_projection(dset):
x, y = dset['lon'].values, dset['lat'].values
m = Basemap(projection='mill', llcrnrlon=dset['lon'].min()-1., llcrnrlat=dset['lat'].min()-1.,
urcrnrlon=dset['lon'].max()+1., urcrnrlat=dset['lat'].max()+1., resolution='i')
m.drawcountries(linewidth=0.5, color='black')
m.shadedrelief()
m.drawcoastlines()
m.drawparallels(np.arange(-90.0, 90.0, 5.), linewidth=0.5, color='white',
labels=[True, False, False, True], fontsize=7)
m.drawmeridians(np.arange(0.0, 360.0, 5.), linewidth=0.5, color='white',
labels=[True, False, False, True], fontsize=7)
x, y = m(x, y)
return(m, x, y)
def truncate_colormap(cmap, minval=0.0, maxval=1.0, n=256):
"""Truncate a colormap by specifying the start and endpoint."""
new_cmap = colors.LinearSegmentedColormap.from_list(
'trunc({n},{a:.2f},{b:.2f})'.format(n=cmap.name, a=minval, b=maxval),
cmap(np.linspace(minval, maxval, n)))
return(new_cmap)
fig = plt.figure(figsize=(10,10))
m, x, y = get_projection(dset)
cmap = truncate_colormap(plt.get_cmap('terrain'), 0.2, 1.)
m.contourf(x, y, dset['topography_c'], tri=True, cmap=cmap, levels=np.arange(-100., 3000., 10.))
plt.show(block=True)
Initial and boundary conditions
Initial and boundary conditions are needed to run any simulation that is not idealized. They contain the main prognostic variables needed by ICON to initialize and run a simulation. These have to be remapped to the horizontal grid created in the first step before running the simulation. Vertical interpolation, instead, is done at runtime inside ICON. Note that there are different initialization mode depending on the input variables. In the following, I'll describe one of the easiest modes which uses only the most important variables.
Input data to generate initial and boundary conditions can be provided by different models, for example ECMWF-IFS, COSMO and ICON itself. In general downloading COSMO and ICON data is easier as it only requires an account on pamore, the system used at DWD to store model output data (see here https://www.dwd.de/DE/leistungen/pamore/pamore.html). Once gained access to the portal (https://webservice.dwd.de/cgi-bin/spp1167/webservice.cgi) data from the operational models used at DWD can be downloaded and then processed. For example, to request ICON-EU data to run a simulation one should use the following command on pamore
pamore -G -F -d 2017090912 -de 2017091012 -tflag best -ee hhl%genv,t%genv,u%genv,v%genv,w%genv,qv%genv,qc%genv,qi%genv,qr%genv,qs%genv,p%genv,t_g,t_ice,h_ice,qv_s,freshsnw,w_snow,t_snow,w_i,rho_snow,t_so,w_so -ires r3b08_n02 -lt m -hindcast_ilam -model ieu
In order to follow along this tutorial a set of ICON-EU data downloaded from pamore can be found here /pool/data/ICON/input_files.tar.gz. Download and untar the file and you will find one GRIB file for every hour with all the necessary variables inside (I have already included the HHL variable in every one of this file but remember to do that if you download new files!).
Create the initial condition
The process of generating the initial condition simply consists in taking the input data (see paragraph before) and interpolating to the target grid that will be used in the simulation (in our case the grids italy_grid_nested_1231m.nc, italy_grid_nested_616m.nc and italy_grid_nested_308m.nc). In theory, this could be done also using CDO or other software but the icontools contain some powerful remapping utilities and convert the input file already to a format that ICON can understand. Furthermore, there are scripts already prepared to do that.
You need an initial condition file for every domain of your simulation! Otherwise, another option is to create an initial condition only for the outermost domain (this is always necessary) and delay the initialization of the other inner nests using the namelist parameter &grid_nml, start_time when running the simulation.
The minimal variables that need to be in the initial condition are
| Variable | Name |
| U | Horizontal Wind Component |
| V | Horizontal Wind Component |
| W | Vertical Wind Component |
| T | Temperature |
| QV | Water Vapour mixing ratio |
| QC | Cloud Water Content |
| QI | Cloud Ice Content |
| P | Pressure |
| QV_S | Surface specific humidity |
| W_SO | Soil Moisture |
| T_SO | Soil Temperature |
| W_SNOW | Snow Water Equivalent |
| RHO_SNOW | Snow density |
| T_G | Surface temperature |
| freshsnow | Age of snow indicator |
| W_I | Snow temperature |
| T_ICE | Sea ice temperature |
| H_ICE | Sea ice depth |
| HHL | Vertical levels |
The script needed to remap the data depends on the input data and is different in case of ECMWF-IFS or ICON data. In this tutorial, we'll use data from ICON-EU. In order to create the initial condition, one needs to use the script mistral_remap_inidata located in the icontools repository. Here is a slightly different version where instead of looping over the variables we define every variable explicitly in order to be able to change their name. Note that you'll need to modify the modules load at the beginning to comply with the ones that you used to compile the icontools.
- xce_remap_inidata.sh
#! /bin/bash
#
# Adapted version of xce_remap_inidata for Mistral
# Created by Guido Cioni (guido.cioni@mpimet.mpg.de)
#-----------------------------------------------------------------------------
#SBATCH --account=bm0682
#SBATCH --job-name=inidata.run
#SBATCH --partition=compute2
#SBATCH --nodes=1
#SBATCH --threads-per-core=2
#SBATCH --output=LOG.inidata.run.%j.o
#SBATCH --error=LOG.inidata.run.%j.o
#SBATCH --exclusive
#SBATCH --time=00:30:00
#=============================================================================
set -x
#---------------------------------------------------------------------------
# openmp environment variables
# ----------------------------
export OMP_NUM_THREADS=6 # 6 on compute2
export ICON_THREADS=6 # 6 on compute2
export OMP_SCHEDULE=dynamic,1
export OMP_DYNAMIC="false"
export OMP_STACKSIZE=4096M
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/sw/rhel6-x64/netcdf/netcdf_fortran-4.4.3-gcc62/lib/
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/sw/rhel6-x64/netcdf/netcdf_c-4.4.1.1-gcc48/lib
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/sw/rhel6-x64/hdf5/hdf5-1.8.14-gcc48/lib
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/sw/rhel6-x64/eccodes/eccodes-2.5.0-gcc48/lib/
#-----------------------------------------------------------------------------
# MPI variables
# ----------------------------
mpi_root=/sw/rhel6-x64/intel/impi/5.0.3.048/lib64
no_of_nodes=${SLURM_JOB_NUM_NODES:-1}
mpi_procs_pernode=6 # 6 or 12 (hyperthreading) on compute2
mpi_total_procs=$((no_of_nodes * mpi_procs_pernode))
START="srun --kill-on-bad-exit=1 --nodes=${SLURM_JOB_NUM_NODES:-1} --ntasks-per-node=${mpi_procs_pernode}"
# load modules
module purge
module load intel gcc
module list
ulimit -s unlimited
# SETTINGS: DIRECTORIES AND INPUT/OUTPUT FILE NAMES --------------------------
# directory containing DWD icon tools binaries
ICONTOOLS_DIR=/work/mh0731/m300382/dwd_icon_tools_git/icontools
# file name of input grid
INGRID=/work/mh0731/m300382/icon_grid_0027_R03B08_N02.nc
# file name of limited-area (output) grid
LOCALGRID=/work/mh0731/m300382/test_nwp_new/italy_grid_nested_1231m.nc
# directory containing data files which shall be mapped to limited-area grid
DATADIR=/work/mh0731/m300382/test_nwp/data
DATAFILELIST=$(find ${DATADIR}/iefff00000000)
# output directory for extracted boundary data
OUTDIR=/work/mh0731/m300382/test_nwp_new/
mkdir -p $OUTDIR
#-----------------------------------------------------------------------------
BINARY_ICONSUB=iconsub_mpi
BINARY_REMAP=iconremap_mpi
AUXGRID=auxgrid
#-----------------------------------------------------------------------------
# Remap inital data onto local (limited-area) grid
#-----------------------------------------------------------------------------
cd ${OUTDIR}
set +x
cat >> NAMELIST_ICONREMAP_FIELDS << EOF_2A
&input_field_nml
inputname = "u"
outputname = "U"
intp_method = 3
/
&input_field_nml
inputname = "v"
outputname = "V"
intp_method = 3
/
&input_field_nml
inputname = "wz"
outputname = "W"
intp_method = 3
/
&input_field_nml
inputname = "t"
outputname = "T"
intp_method = 3
/
&input_field_nml
inputname = "q"
outputname = "QV"
intp_method = 3
/
&input_field_nml
inputname = "clwmr"
outputname = "QC"
intp_method = 3
/
&input_field_nml
inputname = "QI"
outputname = "QI"
intp_method = 3
/
&input_field_nml
inputname = "rwmr"
outputname = "QR"
intp_method = 3
/
&input_field_nml
inputname = "snmr"
outputname = "QS"
intp_method = 3
/
&input_field_nml
inputname = "pres"
outputname = "P"
intp_method = 3
/
&input_field_nml
inputname = "W_SO"
outputname = "W_SO"
intp_method = 3
/
&input_field_nml
inputname = "T_SO"
outputname = "T_SO"
intp_method = 3
/
&input_field_nml
inputname = "sd"
outputname = "W_SNOW"
intp_method = 3
/
&input_field_nml
inputname = "rsn"
outputname = "RHO_SNOW"
intp_method = 3
/
&input_field_nml
inputname = "T_SNOW"
outputname = "T_SNOW"
intp_method = 3
/
&input_field_nml
inputname = "QV_S"
outputname = "QV_S"
intp_method = 3
/
&input_field_nml
inputname = "T_G"
outputname = "T_G"
intp_method = 3
/
&input_field_nml
inputname = "FRESHSNW"
outputname = "freshsnow"
intp_method = 3
/
&input_field_nml
inputname = "cnwat"
outputname = "W_I"
intp_method = 3
/
&input_field_nml
inputname = "icetk"
outputname = "H_ICE"
intp_method = 3
/
&input_field_nml
inputname = "ist"
outputname = "T_ICE"
intp_method = 3
/
&input_field_nml
inputname = "HHL"
outputname = "HHL"
intp_method = 3
/
EOF_2A
set -x
cat NAMELIST_ICONREMAP_FIELDS
#-----------------------------------------------------------------------------
# loop over file list:
echo ${DATAFILELIST}
for datafilename in ${DATAFILELIST} ; do
datafile="${datafilename##*/}" # get filename without path
outdatafile=${datafile%.*} # get filename without suffix
cat > NAMELIST_ICONREMAP << EOF_2B
&remap_nml
in_grid_filename = '${INGRID}'
in_filename = '${DATADIR}/${datafile}'
in_type = 2
out_grid_filename = '${LOCALGRID}'
out_filename = '${OUTDIR}/${outdatafile}.nc'
out_type = 2
out_filetype = 4
l_have3dbuffer = .false.
ncstorage_file = "ncstorage.tmp"
/
EOF_2B
${START} ${ICONTOOLS_DIR}/${BINARY_REMAP} \
--remap_nml NAMELIST_ICONREMAP \
--input_field_nml NAMELIST_ICONREMAP_FIELDS 2>&1
done
#-----------------------------------------------------------------------------
# clean-up
rm -f ncstorage.tmp*
rm -f nml.log NAMELIST_SUB NAMELIST_ICONREMAP NAMELIST_ICONREMAP_FIELDS
#-----------------------------------------------------------------------------
exit
#-----------------------------------------------------------------------------
For more information about the options have a look a the icontools guide https://code.mpimet.mpg.de/projects/dwd-icon-tools/repository/revisions/master/entry/doc/icontools_doc.pdf. The most important parameters to modify are
ICONTOOLS_DIR= # directory containing DWD icon tools binaries
INGRID=# file name of input grid
LOCALGRID= # file name of limited-area (output) grid
DATADIR= # directory containing data files which shall be mapped to limited-area grid
OUTDIR= # output directory for extracted boundary data
Please note the difference between INGRID and OUTDIR. Why is the first one required? Input data, as the one that I'm providing in the example, do not come with grid information to save space. For this reason, icontools need to know where the original grid is. You have to download the appropriate grid for the data. In the case of ICON go on the DWD website where all the grids are stored and find the one that is used in the operational version of ICON. Once you download the grid file make sure that the number of points is the same as in the input data: this way you are pretty sure that the grid has to be the same.
Send the code to the queue, a new netcdf file should be created in the directory provided (iefff00000000.nc). Try to open the file with Paraview and check if everything looks physically sound :). If you get segmentation fault during the interpolation try to change the method; RBF doesn't always work as expected. Always refers to the icontools official documentation and contact Florian Prill at DWD if you still have problems.
GRIB is a moving target. Sometimes if the parameter tables are not correctly loaded in your system you'll have problem running the script as icontools won't be able to recognize the variale names in the grib file. So make sure beforehand that you can load the correct name by checking with cdo sinfov on the file. Otherwise you can even convert the files to netcdf before and use them in icontools.
Small digression on IFS input data
In theory when using data from the ECMWF-IFS model as input one has only to modify the set of variables needed as input and the in_type parameter in the remap_nml. Here is an example (not up to date!):
- remap_inidata.run
#! /bin/bash
#
# Adapted version of xce_remap_inidata for Mistral
# Created by Guido Cioni (guido.cioni@mpimet.mpg.de)
#-----------------------------------------------------------------------------
#SBATCH --account=bm0682
#SBATCH --job-name=inidata.run
#SBATCH --partition=compute2
#SBATCH --nodes=1
#SBATCH --threads-per-core=2
#SBATCH --output=LOG.inidata.run.%j.o
#SBATCH --error=LOG.inidata.run.%j.o
#SBATCH --exclusive
#SBATCH --time=00:30:00
#=============================================================================
set -x
#---------------------------------------------------------------------------
# openmp environment variables
# ----------------------------
export OMP_NUM_THREADS=6 # 6 on compute2
export ICON_THREADS=6 # 6 on compute2
export OMP_SCHEDULE=dynamic,1
export OMP_DYNAMIC="false"
export OMP_STACKSIZE=4096M
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/sw/rhel6-x64/netcdf/netcdf_fortran-4.4.3-gcc62/lib/
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/sw/rhel6-x64/netcdf/netcdf_c-4.4.1.1-gcc48/lib
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/sw/rhel6-x64/hdf5/hdf5-1.8.14-gcc48/lib
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/sw/rhel6-x64/grib_api/grib_api-1.15.0-gcc48/lib
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/sw/rhel6-x64/eccodes/eccodes-2.5.0-gcc48/lib/
#-----------------------------------------------------------------------------
# MPI variables
# ----------------------------
mpi_root=/sw/rhel6-x64/intel/impi/5.0.3.048/lib64
no_of_nodes=${SLURM_JOB_NUM_NODES:-1}
mpi_procs_pernode=6 # 6 or 12 (hyperthreading) on compute2
mpi_total_procs=$((no_of_nodes * mpi_procs_pernode))
START="srun --kill-on-bad-exit=1 --nodes=${SLURM_JOB_NUM_NODES:-1} --ntasks-per-node=${mpi_procs_pernode}"
# load modules
module purge
module load intel gcc
module list
ulimit -s unlimited
# SETTINGS: DIRECTORIES AND INPUT/OUTPUT FILE NAMES --------------------------
# directory containing DWD icon tools binaries
ICONTOOLS_DIR=/work/mh0731/m300382/dwd_icon_tools_git/icontools
# file name of input grid
INGRID=/work/mh0731/m300382/mesovortices/data/ifs_oper_T1279_2014022400.grb
# file name of limited-area (output) grid
LOCALGRID=/work/mh0731/m300382/mesovortices/grid_NA_2492m.nc
# directory containing data files which shall be mapped to limited-area grid
DATADIR=/work/mh0731/m300382/mesovortices/data/
DATAFILELIST=$(find ${DATADIR}/ifs_oper_T1279_2014022400.grb.nc)
# output directory for extracted boundary data
OUTDIR=/work/mh0731/m300382/mesovortices/
mkdir -p $OUTDIR
#-----------------------------------------------------------------------------
BINARY_ICONSUB=iconsub_mpi
BINARY_REMAP=iconremap_mpi
AUXGRID=auxgrid
#-----------------------------------------------------------------------------
# Remap inital data onto local (limited-area) grid
#-----------------------------------------------------------------------------
cd ${OUTDIR}
set +x
cat >> NAMELIST_ICONREMAP_FIELDS << EOF_2A
&input_field_nml
inputname = "u"
outputname = "U"
intp_method = 3
/
&input_field_nml
inputname = "v"
outputname = "V"
intp_method = 3
/
&input_field_nml
inputname = "w"
outputname = "W"
intp_method = 3
/
&input_field_nml
inputname = "t"
outputname = "T"
intp_method = 3
/
&input_field_nml
inputname = "q"
outputname = "QV"
intp_method = 3
/
&input_field_nml
inputname = "clwc"
outputname = "QC"
intp_method = 3
/
&input_field_nml
inputname = "ciwc"
outputname = "QI"
intp_method = 3
/
&input_field_nml
inputname = "crwc"
outputname = "QR"
intp_method = 3
/
&input_field_nml
inputname = "cswc"
outputname = "QS"
intp_method = 3
/
&input_field_nml
inputname = "z_2"
outputname = "GEOP_SFC"
intp_method = 3
/
&input_field_nml
inputname = "z"
outputname = "GEOP_ML"
intp_method = 3
/
&input_field_nml
inputname = "tsn"
outputname = "T_SNOW"
intp_method = 3
/
&input_field_nml
inputname = "sd"
outputname = "W_SNOW"
intp_method = 3
/
&input_field_nml
inputname = "rsn"
outputname = "RHO_SNOW"
intp_method = 3
/
&input_field_nml
inputname = "asn"
outputname = "ALB_SNOW"
intp_method = 3
/
&input_field_nml
inputname = "skt"
outputname = "SKT"
intp_method = 3
/
&input_field_nml
inputname = "sst"
outputname = "SST"
intp_method = 3
/
&input_field_nml
inputname = "stl1"
outputname = "STL1"
intp_method = 3
/
&input_field_nml
inputname = "stl2"
outputname = "STL2"
intp_method = 3
/
&input_field_nml
inputname = "stl3"
outputname = "STL3"
intp_method = 3
/
&input_field_nml
inputname = "stl4"
outputname = "STL4"
intp_method = 3
/
&input_field_nml
inputname = "swvl1"
outputname = "SMIL1"
intp_method = 3
/
&input_field_nml
inputname = "swvl2"
outputname = "SMIL2"
intp_method = 3
/
&input_field_nml
inputname = "swvl3"
outputname = "SMIL3"
intp_method = 3
/
&input_field_nml
inputname = "swvl4"
outputname = "SMIL4"
intp_method = 3
/
&input_field_nml
inputname = "ci"
outputname = "CI"
intp_method = 3
/
&input_field_nml
inputname = "src"
outputname = "W_I"
intp_method = 3
/
&input_field_nml
inputname = "SR"
outputname = "Z0"
intp_method = 3
/
&input_field_nml
inputname = "lsm"
outputname = "LSM"
intp_method = 3
/
&input_field_nml
inputname = "lnsp"
outputname = "LNPS"
intp_method = 3
/
EOF_2A
set -x
cat NAMELIST_ICONREMAP_FIELDS
#-----------------------------------------------------------------------------
# loop over file list:
echo ${DATAFILELIST}
for datafilename in ${DATAFILELIST} ; do
datafile="${datafilename##*/}" # get filename without path
outdatafile=${datafile%.*} # get filename without suffix
cat > NAMELIST_ICONREMAP << EOF_2B
&remap_nml
in_grid_filename = '${INGRID}'
in_filename = '${DATADIR}/${datafile}'
in_type = 1
out_grid_filename = '${LOCALGRID}'
out_filename = '${OUTDIR}/${outdatafile}.nc'
out_type = 2
out_filetype = 5
l_have3dbuffer = .false.
ncstorage_file = "ncstorage.tmp"
/
EOF_2B
${START} ${ICONTOOLS_DIR}/${BINARY_REMAP} \
--remap_nml NAMELIST_ICONREMAP \
--input_field_nml NAMELIST_ICONREMAP_FIELDS 2>&1
done
#-----------------------------------------------------------------------------
# clean-up
rm -f ncstorage.tmp*
rm -f nml.log NAMELIST_SUB NAMELIST_ICONREMAP NAMELIST_ICONREMAP_FIELDS
#-----------------------------------------------------------------------------
exit
#-----------------------------------------------------------------------------
Small digression on COSMO input data
The set of variables coming from the COSMO model needed to initialize ICON are slightly different. Here is an example
- run_inidata_cosmo.sh
#!/bin/bash
# ===== SLURM settings ========================================================
#SBATCH --account=bm0682
#SBATCH --job-name=inidata.run
#SBATCH --partition=compute2
#SBATCH --nodes=1
#SBATCH --threads-per-core=2
#SBATCH --output=LOG.inidata.run.%j.o
#SBATCH --error=LOG.inidata.run.%j.o
#SBATCH --exclusive
#SBATCH --time=00:30:00
#=============================================================================
set -x
#---------------------------------------------------------------------------
# openmp environment variables
# ----------------------------
export OMP_NUM_THREADS=6
export ICON_THREADS=6
export OMP_SCHEDULE=dynamic,1
export OMP_DYNAMIC="false"
export OMP_STACKSIZE=4096M
#-----------------------------------------------------------------------------
# MPI variables
# ----------------------------
mpi_root=/sw/rhel6-x64/intel/impi/5.0.3.048/lib64
no_of_nodes=${SLURM_JOB_NUM_NODES:-1}
mpi_procs_pernode=6
mpi_total_procs=$((no_of_nodes * mpi_procs_pernode))
START="srun --kill-on-bad-exit=1 --nodes=${SLURM_JOB_NUM_NODES:-1} --ntasks-per-node=${mpi_procs_pernode}"
# load modules
module purge
# module load intelmpi/2018.0.128 intel/18.0.0 gcc/7.1.0
module load intel gcc
module list
ulimit -s unlimited
# ===== load other modules ====================================================
module load nco
module load cdo
#=============================================================================
# ===== directory and file name settings ======================================
# all names of intermediate, temporary files contain ${SLURM_JOB_ID} to avoid
# accidentally overwriting the intermediate files of another job
BINDIR=/work/mh0731/m300382/dwd_icon_tools_git/icontools
SCRATCHDIR=/scratch/m/m300382
OUTDIR=/work/mh0731/m300382/tornado_dolo
# INPUT FILE
INFILE_COSMO=/work/mh0731/m300382/tornado_dolo/pamore/smi/laf2015070806.nc
# OUTPUT FILE
# /work/bm0834/k203095/pool/GRIDS/HDCP_FINAL_4/hdcp2_cosmodom_R0625m.nc
# /work/bm0834/k203095/pool/GRIDS/HDCP_FINAL_4/hdcp2_cosmodom_nest_R0312m.nc
# /work/bm0834/k203095/pool/GRIDS/HDCP_FINAL_4/hdcp2_cosmodom_nest_R0156m.nc
OUTGRIDFILE=/work/mh0731/m300382/tornado_dolo/tornado_parent0622m.nc
OUTFILE=${OUTDIR}/init_tornado_parent622m_2015070806.nc
# ====== check files ==========================================================
for file in ${INFILE_COSMO} ${OUTGRIDFILE}; do
if [ ! -f ${file} ]; then
echo "${file} not found. Exiting..."
exit 1
fi
done
i=0
while [ -f ${OUTFILE} ]; do
if [ $i -eq 0 ]; then
OUTFILE=${OUTFILE}_00
fi
i=$((i+1))
echo "${OUTFILE} exists. Saving results to ${OUTFILE}_$(printf '%02d' $i)"
OUTFILE=${OUTFILE%??}$(printf '%02d' $i)
done
# ====== make directories =====================================================
mkdir -p ${SCRATCHDIR}
mkdir -p ${OUTDIR}
# ====== interpolation methods, grid types, and file types ====================
# set these individually for each variable in the namelists below
CONS=2 #conservative
RBF=3 #radial basis funcion
NN=4 #nearest neighbour
intp_method=3
GLOBREG=1
ICONTRI=2
LOCREG=3
GRIB=2
NC2=4
NC4=5
# ====== remap constant HHL field =============================================
echo "Processing ${INFILE_COSMO}."
cat > ${SCRATCHDIR}/remap_init_HHL_${SLURM_JOB_ID}.nml << EOF
! REMAPPING NAMELIST FILE
!
&remap_nml
in_grid_filename = "${INFILE_COSMO}"
in_filename = "${INFILE_COSMO}"
in_type = ${LOCREG}
!
out_grid_filename = "${OUTGRIDFILE}"
out_filename = "${SCRATCHDIR}/lffd_remap_HHL_${SLURM_JOB_ID}.nc"
out_type = ${ICONTRI}
!
out_filetype = ${NC4} !use NetCDF Classic because ncrename does not work with NetCDF 4
/
! DEFINITION FOR INPUT DATA FIELD
!
&input_field_nml
inputname = "HHL"
outputname = "HHL"
intp_method = ${RBF}
/
EOF
${srun} ${BINDIR}/iconremap_mpi --remap_nml ${SCRATCHDIR}/remap_init_HHL_${SLURM_JOB_ID}.nml
# Rename variable and dimensions according to ICON definition
#ncrename -d height,lev_2 -v height,lev_2 "${SCRATCHDIR}/lffd_remap_HHL_${SLURM_JOB_ID}.nc"
#ncrename -d height,lev_3 -v height,lev_3 "${SCRATCHDIR}/lffd_remap_HHL_${SLURM_JOB_ID}.nc"
# Add to outputfile
#ncks -A -v HHL ${SCRATCHDIR}/lffd_remap_HHL_${SLURM_JOB_ID}.nc ${OUTFILE}
# ====== remap COSMO-DE fields ================================================
# Exclude U and V wind components because of different coordinates,
# which are a problem for the dwdtools
UVARS=U,AUMFL_S,srlon,slonu,slatu
VVARS=V,AVMFL_S,srlat,slonv,slatv
ncks -O -x -v ${UVARS},${VVARS} ${INFILE_COSMO} ${SCRATCHDIR}/lffd_xUV_${SLURM_JOB_ID}.nc
echo "Processing ${INFILE_COSMO}."
echo "Processing ${SCRATCHDIR}/lffd_xUV_${SLURM_JOB_ID}.nc."
cat > ${SCRATCHDIR}/remap_init_${SLURM_JOB_ID}.nml << EOF
! REMAPPING NAMELIST FILE
!
&remap_nml
in_grid_filename = "${INFILE_COSMO}"
in_filename = "${SCRATCHDIR}/lffd_xUV_${SLURM_JOB_ID}.nc"
in_type = ${LOCREG}
!
out_grid_filename = "${OUTGRIDFILE}"
out_filename = "${SCRATCHDIR}/lffd_remap_xUV_${SLURM_JOB_ID}.nc"
out_type = ${ICONTRI}
!
out_filetype = ${NC4}
/
! DEFINITION FOR INPUT DATA FIELD
!
&input_field_nml
inputname = "T"
outputname = "T"
intp_method = ${intp_method}
/
&input_field_nml
inputname = "QC"
outputname = "QC"
intp_method = ${intp_method}
/
&input_field_nml
inputname = "W"
outputname = "W"
intp_method = ${intp_method}
/
&input_field_nml ! geopotential
inputname = "QR"
outputname = "QR"
intp_method = ${intp_method}
/
&input_field_nml ! geopotential
inputname = "QS"
outputname = "QS"
intp_method = ${intp_method}
/
&input_field_nml ! geopotential
inputname = "QV"
outputname = "QV"
intp_method = ${intp_method}
/
&input_field_nml ! geopotential
inputname = "QI"
outputname = "QI"
intp_method = ${intp_method}
/
&input_field_nml ! geopotential
inputname = "PS"
outputname = "PS"
intp_method = ${intp_method}
/
&input_field_nml ! geopotential
inputname = "P"
outputname = "P"
intp_method = ${intp_method}
/
&input_field_nml ! ist FIS
inputname = "FIS"
outputname = "GEOSP"
intp_method = ${intp_method}
/
&input_field_nml ! Plant Canopy Surface Water
inputname = "W_I"
outputname = "w_i"
intp_method = ${intp_method}
/
&input_field_nml ! Snow depth water equivalent
inputname = "W_SNOW"
outputname = "w_snow"
intp_method = ${intp_method}
/
&input_field_nml ! horiz. wind comp. v
inputname = "T_G"
outputname = "t_g"
intp_method = ${intp_method}
/
&input_field_nml ! vertical velocity
inputname = "T_SNOW"
outputname = "t_snow"
intp_method = ${intp_method}
/
&input_field_nml ! surface pressure
inputname = "T_ICE"
outputname = "t_ice"
intp_method = ${intp_method}
/
&input_field_nml ! geopotential
inputname = "QV_S"
outputname = "qv_s"
intp_method = ${intp_method}
/
&input_field_nml ! geopotential
inputname = "H_SNOW"
outputname = "h_snow"
intp_method = ${intp_method}
/
&input_field_nml ! geopotential
inputname = "H_ICE"
outputname = "h_ice"
intp_method = ${intp_method}
/
&input_field_nml ! geopotential
inputname = "FRESHSNW"
outputname = "freshsnow"
intp_method = ${intp_method}
/
&input_field_nml ! geopotential
inputname = "Z0"
outputname = "gz0"
intp_method = ${intp_method}
/
&input_field_nml ! soil ice content (multilayers)
inputname = "W_SO_ICE"
outputname = "w_so_ice"
intp_method = ${intp_method}
/
&input_field_nml ! Column-integrated Soil Moisture (multilayers)
inputname = "SMI"
outputname = "smi"
intp_method = 4
/
&input_field_nml ! Soil Temperature (multilayers)
inputname = "T_SO"
outputname = "t_so"
intp_method = 4
/
&input_field_nml ! geopotential
inputname = "RHO_SNOW"
outputname = "rho_snow"
intp_method = ${intp_method}
/
EOF
${srun} ${BINDIR}/iconremap_mpi --remap_nml ${SCRATCHDIR}/remap_init_${SLURM_JOB_ID}.nml
### copy .nc4 to .nc2
#cdo -f nc2 copy ${SCRATCHDIR}/lffd_remap_xUV_${SLURM_JOB_ID}.nc4 ${SCRATCHDIR}/lffd_remap_xUV_${SLURM_JOB_ID}.nc
#Error (cdf_enddef) : NetCDF: One or more variable sizes violate format constraints
#Error (cdf_close) : NetCDF: HDF error
# Rename variable and dimensions according to ICON definition
#ncrename -d height,lev -d height_2,lev_2 -v height,lev -v height_2,lev_2 ${SCRATCHDIR}/lffd_remap_xUV_${SLURM_JOB_ID}.nc
# Add to outputfile
#ncks -A -v T,W,P,PS,GEOSP,QV,QC,QI,QR,QS,w_i,w_snow,t_g,t_snow,t_ice,qv_s,h_snow,h_ice,freshsnow,gz0,w_so_ice,smi,t_so,rho_snow ${SCRATCHDIR}/lffd_remap_xUV_${SLURM_JOB_ID}.nc ${OUTFILE}
##ncks -A -v height,height_2 ${INFILE_COSMO} ${OUTFILE}
# ====== remap extracted wind fields from COSMO file ==========================
# remapping the U and V wind fields with the other fields does not work because
# the DWD ICON Tools cannot handle multiple horizontal grids in one file
for var in U V; do
ncks -O -v ${var} ${INFILE_COSMO} ${SCRATCHDIR}/lffd_${var}_${SLURM_JOB_ID}.nc
echo "Processing ${SCRATCHDIR}/lffd_${var}_${SLURM_JOB_ID}.nc."
cat > ${SCRATCHDIR}/remap_init_${SLURM_JOB_ID}.nml << EOF
! REMAPPING NAMELIST FILE
!
&remap_nml
in_grid_filename = "${INFILE_COSMO}"
in_filename = "${SCRATCHDIR}/lffd_${var}_${SLURM_JOB_ID}.nc"
in_type = ${LOCREG}
!
out_grid_filename = "${OUTGRIDFILE}"
out_filename = "${SCRATCHDIR}/lffd_remap_${var}_${SLURM_JOB_ID}.nc"
out_type = ${ICONTRI}
!
out_filetype = ${NC4}
/
! DEFINITION FOR INPUT DATA FIELD
&input_field_nml
inputname = "${var}"
outputname = "${var}"
intp_method = ${RBF}
/
EOF
${srun} ${BINDIR}/iconremap_mpi --remap_nml ${SCRATCHDIR}/remap_init_${SLURM_JOB_ID}.nml
# Rename variable and dimensions according to ICON definition
# ncrename -d height,lev -v height,lev ${SCRATCHDIR}/lffd_remap_${var}_${SLURM_JOB_ID}.nc
# Add to outputfile
#ncks -A -v ${var} ${SCRATCHDIR}/lffd_remap_${var}_${SLURM_JOB_ID}.nc ${OUTFILE}
done
# ====== Merge ==========================
cdo -P 16 -O merge ${SCRATCHDIR}/lffd_remap_xUV_${SLURM_JOB_ID}.nc ${SCRATCHDIR}/lffd_remap_[UV]_${SLURM_JOB_ID}.nc ${SCRATCHDIR}/lffd_remap_HHL_${SLURM_JOB_ID}.nc ${OUTFILE}
cdo sinfov ${OUTFILE}
# Average over bnds dimension, to delete this unused dimension
ncwa -O -a bnds -d bnds,1,1 ${OUTFILE} ${OUTFILE}
cdo sinfov ${OUTFILE}
pwd
Create boundary conditions
The procedure to create the boundary conditions is similar only that this time you have less variables to interpolate but more files (can be one every hour, 3 or 6 hours depending on the setup). In our case it is every hour. The minimal variables that need to be in the boundary conditions are
Boundary conditions variables
Boundary conditions variables
| Variable | Name |
| U | Horizontal Wind Component |
| V | Horizontal Wind Component |
| W | Vertical Wind Component |
| T | Temperature |
| QV | Water Vapour mixing ratio |
| QC | Cloud Water Content |
| QI | Cloud Ice Content |
| P | Pressure |
| HHL | Vertical levels |
In the newest version of ICON the possibility exists to create an additional grid defined only over the boundary of the domain where boundary conditions are prescribed. This grid is then used for the remapping, thus saving space in the output files. Until now boundary conditions were remapped to the same grid as the initial condition but this was (1) wasting space and (2) causing slowdown at runtime due to the reading of unnecessary data. Users should now use this different method which is explained hereinafter. The only difference with the old method is that the binary iconsub in the icontools is used to create the grid lateral_boundary.grid.nc which is then used to remap
The relevant script is
Boundary Conditions generation
Boundary Conditions generation
#! /bin/bash
#-----------------------------------------------------------------------------
#SBATCH --account=bm0682
#SBATCH --job-name=latbc.run
#SBATCH --partition=compute2
#SBATCH --nodes=1
#SBATCH --threads-per-core=2
#SBATCH --output=LOG.latbc.run.%j.o
#SBATCH --error=LOG.latbc.run.%j.o
#SBATCH --exclusive
# #
#SBATCH --time=2:00:00
#=============================================================================
set -x
#---------------------------------------------------------------------------
# openmp environment variables
# ----------------------------
export OMP_NUM_THREADS=6
export ICON_THREADS=6
export OMP_SCHEDULE=dynamic,1
export OMP_DYNAMIC="false"
export OMP_STACKSIZE=4096M
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/sw/rhel6-x64/netcdf/netcdf_fortran-4.4.3-gcc62/lib/
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/sw/rhel6-x64/netcdf/netcdf_c-4.4.1.1-gcc48/lib
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/sw/rhel6-x64/hdf5/hdf5-1.8.14-gcc48/lib
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/sw/rhel6-x64/eccodes/eccodes-2.5.0-gcc48/lib/
#-----------------------------------------------------------------------------
# MPI variables
# ----------------------------
mpi_root=/sw/rhel6-x64/intel/impi/5.0.3.048/lib64
no_of_nodes=${SLURM_JOB_NUM_NODES:-1}
mpi_procs_pernode=6
mpi_total_procs=$((no_of_nodes * mpi_procs_pernode))
START="srun --kill-on-bad-exit=1 --nodes=${SLURM_JOB_NUM_NODES:-1} --ntasks-per-node=${mpi_procs_pernode}"
# load modules
module purge
module load intelmpi/5.0.3.048 intel git
module list
ulimit -s unlimited
# SETTINGS: DIRECTORIES AND INPUT/OUTPUT FILE NAMES --------------------------
# directory containing DWD icon tools binaries
ICONTOOLS_DIR=/work/mh0731/m300382/dwd_icon_tools_git/icontools
# file name of input grid
INGRID=/work/mh0731/m300382/icon_grid_0027_R03B08_N02.nc
# file name of limited-area (output) grid
LOCALGRID=/work/mh0731/m300382/test_nwp_new/italy_grid_nested_1231m.nc
# directory containing data files which shall be mapped to limited-area grid
DATADIR=/work/mh0731/m300382/test_nwp/data
DATAFILELIST=$(find ${DATADIR}/iefff0???0000_2)
# output directory for extracted boundary data
OUTDIR=/work/mh0731/m300382/test_nwp_new
mkdir -p $OUTDIR
#-----------------------------------------------------------------------------
BINARY_ICONSUB=iconsub_mpi
BINARY_REMAP=iconremap_mpi
# grid file defining the lateral boundary
AUXGRID="lateral_boundary"
#-----------------------------------------------------------------------------
# PART I: Create auxiliary grid file which contains only the cells of the
# boundary zone.
#-----------------------------------------------------------------------------
cd ${OUTDIR}
cat > NAMELIST_ICONSUB << EOF_1
&iconsub_nml
grid_filename = '${LOCALGRID}',
output_type = 4,
lwrite_grid = .TRUE.,
/
&subarea_nml
ORDER = "${OUTDIR}/${AUXGRID}",
grf_info_file = '${LOCALGRID}',
min_refin_c_ctrl = 1
max_refin_c_ctrl = 14
/
EOF_1
${START} ${ICONTOOLS_DIR}/${BINARY_ICONSUB} -vvv --nml NAMELIST_ICONSUB 2>&1
#-----------------------------------------------------------------------------
# PART II: Extract boundary data
#-----------------------------------------------------------------------------
#-----------------------------------------------------------------------------
# preparations:
rm -f ncstorage.tmp*
set +x
cat > NAMELIST_ICONREMAP_FIELDS << EOF_2A
&input_field_nml
inputname = "u"
outputname = "u"
intp_method = 3
/
&input_field_nml
inputname = "v"
outputname = "v"
intp_method = 3
/
&input_field_nml
inputname = "wz"
outputname = "w"
intp_method = 3
/
&input_field_nml
inputname = "t"
outputname = "temp"
intp_method = 3
/
&input_field_nml
inputname = "q"
outputname = "qv"
intp_method = 3
/
&input_field_nml
inputname = "clwmr"
outputname = "qc"
intp_method = 3
/
&input_field_nml
inputname = "QI"
outputname = "qi"
intp_method = 3
/
&input_field_nml
inputname = "rwmr"
outputname = "qr"
intp_method = 3
/
&input_field_nml
inputname = "pres"
outputname = "pres"
intp_method = 3
/
&input_field_nml
inputname = "snmr"
outputname = "qs"
intp_method = 3
/
&input_field_nml
inputname = "HHL"
outputname = "z_ifc"
intp_method = 3
/
EOF_2A
set -x
#-----------------------------------------------------------------------------
# loop over file list:
echo ${DATAFILELIST}
for datafilename in ${DATAFILELIST} ; do
datafile="${datafilename##*/}" # get filename without path
outdatafile=${datafile%.*} # get filename without suffix
cat > NAMELIST_ICONREMAP << EOF_2B
&remap_nml
in_grid_filename = '${INGRID}'
in_filename = '${DATADIR}/${datafile}'
in_type = 2
out_grid_filename = '${OUTDIR}/${AUXGRID}.grid.nc'
out_filename = '${OUTDIR}/${outdatafile}_lbc.nc'
out_type = 2
out_filetype = 4
l_have3dbuffer = .false.
ncstorage_file = "ncstorage.tmp"
/
EOF_2B
${START} ${ICONTOOLS_DIR}/${BINARY_REMAP} \
--remap_nml NAMELIST_ICONREMAP \
--input_field_nml NAMELIST_ICONREMAP_FIELDS 2>&1
done
#-----------------------------------------------------------------------------
# clean-up
rm -f ncstorage.tmp*
rm -f nml.log NAMELIST_SUB NAMELIST_ICONREMAP NAMELIST_ICONREMAP_FIELDS
#-----------------------------------------------------------------------------
exit
#-----------------------------------------------------------------------------
Again make sure that all the folders and links in this script are adjusted to your needs before sending the script to the queue.
Every file containing boundary conditions need to have the vertical levels inside. Sometimes this is problematic as data downloaded from pamore only contains the HHL (vertical levels) variable only in the first file, at initialization. Make sure that you copy this variable over to every file using CDO before running the icontools script.
If everything is running smoothly one netcdf file for every hour will be generated, that is iefff00*_2_lbc.nc.
When running the simulation the file name for the boundary conditions will be automatically created using the current simulation date/time. Thus, it is a good idea to rename the boundary conditions files so that they contain the date in the filename. You can easily do that with a bit of bash or python scripting. Here is an example:
- change_file_names.py
import netCDF4
from datetime import datetime
import glob
import os
folder="/work/mh0731/m300382/test_nwp_new/"
#
in_files="iefff0???0000_2_lbc.nc"
#
out_prefix="iefff_"
for filename in sorted(glob.glob(folder+in_files)):
nc_temp = netCDF4.Dataset(filename)
dtime = netCDF4.num2date(nc_temp.variables['time'][:], nc_temp.variables['time'].units)
date_string = datetime.strftime(dtime[0], format='%Y%m%d%H')
print('old = '+filename)
print('new = '+folder+out_prefix+date_string+"_lbc.nc")
os.rename(filename, folder+out_prefix+date_string+"_lbc.nc")
The variables contained in the boundary condition files can vary accordingly to this decision tree, which is taken from /src/io/atmo/mo_async_latbc.f90, ICON will use whatever is present in the file.
!! ------------------------------------------------------------------------
!! Which fields are read from the lateral boundary conditions file?
!! ------------------------------------------------------------------------
!!
!! This question is answered independently from the "init_icon"
!! namelist parameter of the initial state setup!
!!
!! - If "VN" is available, then it is read from file, otherwise "U","V".
!! - "W" is optional and read if available in the input data (note that "W" may in fact contain OMEGA).
!! - "QV", "QC", "QI" are always read
!! - "QR", "QS" are read if available
!!
!! The other fields for the lateral boundary conditions are read
!! from file, based on the following decision tree. Note that the
!! basic distinction between input from non-hydrostatic and
!! hydrostatic models is made on the availability of the HHL field.
!!
!! +------------------+
!! | HHL available? |
!! +------------------+
!! |
!! ______yes_______|________no________
!! | |
!! +--------------------------+ +------------------------+
!! | RHO & THETA_V available? | | PS,GEOP & T available? |
!! +--------------------------+ +------------------------+
!! | |
!! ____yes___|____no______ ___yes__|________no___________
!! | | | |
!! | +----------------+ | +--------+
!! * read HHL,RHO,THETA_V | P,T available? | * read in PS,GEOP,T | ERROR! |
!! * read W if available | | * read OMEGA if available | |
!! * ignore PS,GEOP +----------------+ * compute P,HHL +--------+
!! * diagnose P,T | * CALL OMEGA -> W
!! ___yes__|___no____
!! | |
!! | +--------+
!! * read HHL,P,T | ERROR! |
!! * ignore PS,GEOP +--------+
!! * read W if available
!!
!!
!! Afterwards, we
!! - re-compute the virtual temperature (inside the vertical
!! interpolation subroutine)
!! - (re-)compute RHO (inside the vertical interpolation subroutine)
!! - perform vertical interpolation
Running the simulation
Before running the simulation make sure that your initial/boundary conditions make sense. Most of the crashes can be prevented just by checking the input data before.
Now that we have all the data we can think about running the simulation. In this example we'll attempt to run with all the 3 nested domains at the same time using a similar configuration for all of them. Note that in ICON you can choose a different physical setup for every domain (for example a different number of vertical levels or a different parametrization of convection or turbulence). The setup that we just created is quite expensive to run so remember to use enough nodes (I managed to get it running with 64 nodes). If the nodes are not enough you'll get a segmentation fault but at the end of the LOG file a OOM Killer detected message will appear so that you'll know the memory is not enough.
Here is a sample script to run the simulation for 24 hours using the files that we just generated. In the following we'll go through the most important settings. Of course you'll have to adapt the script to your needs and especially change the modules and libraries loaded at the beginning.
- run_ICON.run
#!/bin/ksh
#=============================================================================
# =====================================
# mistral batch job parameters
#-----------------------------------------------------------------------------
#SBATCH --account=bm0682
#SBATCH --job-name=icon_nwp
#SBATCH --partition=compute2
#SBATCH --workdir=/work/mh0731/m300382/test_nwp_new
#SBATCH --nodes=64
#SBATCH --threads-per-core=2
#SBATCH --output=/work/mh0731/m300382/test_nwp_new/LOG_icon.run.%j.o
#SBATCH --error=/work/mh0731/m300382/test_nwp_new/LOG_icon.run.%j.o
#SBATCH --exclusive
#SBATCH --time=00:15:00
#=============================================================================
#
# ICON run script. Created by ./config/make_target_runscript
# target machine is bullx
# target use_compiler is intel
# with mpi=yes
# with openmp=no
# memory_model=large
# submit with sbatch -N ${SLURM_JOB_NUM_NODES:-1}
#
#=============================================================================
set -x
#. ./add_run_routines
#-----------------------------------------------------------------------------
# target parameters
# ----------------------------
site="dkrz.de"
target="bullx"
compiler="intel"
loadmodule="intel/16.0 ncl/6.2.1-gccsys cdo/default svn/1.8.13 hpcx/1.9.7 openmpi/2.0.2p1_hpcx-intel14"
with_mpi="yes"
with_openmp="no"
job_name="icon_nwp"
submit="sbatch -N ${SLURM_JOB_NUM_NODES:-1}"
#-----------------------------------------------------------------------------
# openmp environment variables
# ----------------------------
export OMP_NUM_THREADS=1
export ICON_THREADS=1
export OMP_SCHEDULE=dynamic,1
export OMP_DYNAMIC="false"
export OMP_STACKSIZE=1024M
#-----------------------------------------------------------------------------
# MPI variables
# ----------------------------
mpi_root=/sw/rhel6-x64/mpi/openmpi-2.0.2p1_hpcx-intel14
no_of_nodes=${SLURM_JOB_NUM_NODES:=1}
mpi_procs_pernode=$((${SLURM_JOB_CPUS_PER_NODE%%\(*} / 2))
((mpi_total_procs=no_of_nodes * mpi_procs_pernode))
START="srun --cpu-freq=HighM1 --kill-on-bad-exit=1 --nodes=${SLURM_JOB_NUM_NODES:-1} --cpu_bind=verbose,cores --distribution=block:block --ntasks=$((no_of_nodes * mpi_procs_pernode)) --ntasks-per-node=${mpi_procs_pernode} --cpus-per-task=$((2 * OMP_NUM_THREADS)) --propagate=STACK,CORE"
#-----------------------------------------------------------------------------
# load ../setting if exists
if [ -a ../setting ]
then
echo "Load Setting"
. ../setting
fi
#-----------------------------------------------------------------------------
bindir="${basedir}/build/x86_64-unknown-linux-gnu/bin" # binaries
BUILDDIR=build/x86_64-unknown-linux-gnu
#-----------------------------------------------------------------------------
#=============================================================================
# load profile
if [ -a /etc/profile ] ; then
. /etc/profile
#=============================================================================
#=============================================================================
# load modules
module purge
module load "$loadmodule"
module list
#=============================================================================
fi
#=============================================================================
export LD_LIBRARY_PATH=/sw/rhel6-x64/netcdf/netcdf_c-4.4.0-parallel-openmpi2-intel14/lib:$LD_LIBRARY_PATH
#=============================================================================
nproma=16
cdo="cdo"
cdo_diff="cdo diffn"
icon_data_rootFolder="/pool/data/ICON"
icon_data_poolFolder="/pool/data/ICON"
icon_data_buildbotFolder="/pool/data/ICON/buildbot_data"
icon_data_buildbotFolder_aes="/pool/data/ICON/buildbot_data/aes"
icon_data_buildbotFolder_oes="/pool/data/ICON/buildbot_data/oes"
export EXPNAME=icon_nwp
export KMP_AFFINITY="verbose,granularity=core,compact,1,1"
export KMP_LIBRARY="turnaround"
export KMP_KMP_SETTINGS="1"
export OMP_WAIT_POLICY="active"
export OMPI_MCA_pml="cm"
export OMPI_MCA_mtl="mxm"
export OMPI_MCA_coll="^fca"
export MXM_RDMA_PORTS="mlx5_0:1"
export HCOLL_MAIN_IB="mlx5_0:1"
export HCOLL_ML_DISABLE_BARRIER="1"
export HCOLL_ML_DISABLE_IBARRIER="1"
export HCOLL_ML_DISABLE_BCAST="1"
export HCOLL_ENABLE_MCAST_ALL="1"
export HCOLL_ENABLE_MCAST="1"
export OMPI_MCA_coll_sync_barrier_after_alltoallv="1"
export OMPI_MCA_coll_sync_barrier_after_alltoallw="1"
export MXM_HANDLE_ERRORS="bt"
export UCX_HANDLE_ERRORS="bt"
export MALLOC_TRIM_THRESHOLD_="-1"
ulimit -s 2097152
ulimit -c 0
# this can not be done in use_mpi_startrun since it depends on the
# environment at time of script execution
#case " $loadmodule " in
# *\ mxm\ *)
# START+=" --export=$(env | sed '/()=/d;/=/{;s/=.*//;b;};d' | tr '\n' ',')LD_PRELOAD=${LD_PRELOAD+$LD_PRELOAD:}${MXM_HOME}/lib/libmxm.so"
# ;;
#esac
#--------------------------------------------------------------------------------------------------
#--------------------------------------------------------------------------------------------------
ICON_BASE_PATH=/work/mh0731/m300382/icon-git
MODEL=${ICON_BASE_PATH}/build/x86_64-unknown-linux-gnu/bin/icon
RUNSCRIPTDIR=${ICON_BASE_PATH}/run
#####################################
EXPDIR=/work/mh0731/m300382/test_nwp_new/exp/test
# the directory for the experiment will be created, if not already there
if [ ! -d $EXPDIR ]; then
mkdir -p $EXPDIR
fi
#
cd $EXPDIR
#
#--------------------------------------------------------------------------------------------------
###### the grid parameters
### Start from 1250 m domain
GRIDFOLDER="/work/mh0731/m300382/test_nwp_new"
grid_1=italy_grid_nested_1231m
grid_2=italy_grid_nested_616m
grid_3=italy_grid_nested_308m
ln -sf ${GRIDFOLDER}/${grid_1}.nc ${EXPDIR}/
ln -sf ${GRIDFOLDER}/${grid_2}.nc ${EXPDIR}/
ln -sf ${GRIDFOLDER}/${grid_3}.nc ${EXPDIR}/
atmo_dyn_grids="${grid_1}.nc ${grid_2}.nc ${grid_3}.nc"
dynamics_parent_grid_id=" 0, 1, 2 "
###### EXT DATA (default filename of external parameter input file, default:”<path>extpar_<gridfile>”)
EXTPARFOLDER=${GRIDFOLDER}
ln -sf ${EXTPARFOLDER}/extpar_${grid_1}.nc ${EXPDIR}/
ln -sf ${EXTPARFOLDER}/extpar_${grid_2}.nc ${EXPDIR}/
ln -sf ${EXTPARFOLDER}/extpar_${grid_3}.nc ${EXPDIR}/
ln -sf ${ICON_BASE_PATH}/data/ECHAM6_CldOptProps.nc rrtm_cldopt.nc
ln -sf ${ICON_BASE_PATH}/data/rrtmg_lw.nc .
ln -sf ${ICON_BASE_PATH}/data/dmin_wetgrowth_lookup.dat .
#--------------------------------------------------------------------------------------------------
#--------------------------------------------------------------------------------------------------
start_date="2018-10-29T00:00:00Z"
end_date="2018-10-30T00:00:00Z"
###### INITIAL DATA
#INIFOLDER=${GRIDFOLDER}
# ln -sf ${INIFOLDER}/iefff00000000.nc ${EXPDIR}/ifs2icon_R2B11_DOM01.nc
# ln -sf ${INIFOLDER}/iefff00000000.nc ${EXPDIR}/dwdFG_R2B11_DOM01.nc
#--------------------------------------------------------------------------------------------------
###### LATBC PATH
# LATBCFOLDER=${GRIDFOLDER}
# ln -sf ${LATBCFOLDER}/lateral_boundary.grid.nc ${EXPDIR}/lateral_boundary.grid.nc
#--------------------------------------------------------------------------------------------------
#--------------------------------------------------------------------------------------------------
#--------------------------------------------------------------------------------------------------
# the namelist filename
atmo_namelist=NAMELIST_${EXPNAME}
#
#
#-----------------------------------------------------------------------------
# model timing
### Start from 1250 m domain
dtime=10
nhours_restart=180
dt_restart=`expr ${nhours_restart} \* 3600 `
nhours_checkpoint=12
dt_checkpoint=`expr ${nhours_checkpoint} \* 3600 `
#
#-----------------------------------------------------------------------------
# model parameters
model_equations=3 # equation system
# 1=hydrost. atm. T
# 1=hydrost. atm. theta dp
# 3=non-hydrost. atm.,
# 0=shallow water model
# -1=hydrost. ocean
#-----------------------------------------------------------------------------
restart=.FALSE.
#-----------------------------------------------------------------------------
# create ICON master namelist
# ------------------------
cat > icon_master.namelist << EOF
&master_nml
lrestart = ${restart}
/
&time_nml
ini_datetime_string = "$start_date"
end_datetime_string = "$end_date"
dt_restart = $dt_restart
/
&master_model_nml
model_type = 1
model_name = "ATMO"
model_namelist_filename = "$atmo_namelist"
model_min_rank = 1
model_max_rank = 65536
model_inc_rank = 1
/
EOF
#-----------------------------------------------------------------------------
#-----------------------------------------------------------------------------
# write ICON namelist parameters
# ------------------------
#
# ------------------------
# reconstrcuct the grid parameters in namelist form
dynamics_grid_filename=""
for gridfile in ${atmo_dyn_grids}; do
dynamics_grid_filename="${dynamics_grid_filename} '${gridfile}'"
done
dynamics_parent_grid_id="${dynamics_parent_grid_id},"
# ------------------------
#
cat > ${atmo_namelist} << EOF
#
¶llel_nml
nproma = 16
p_test_run = .FALSE.
l_test_openmp = .FALSE.
l_log_checks = .FALSE.
num_io_procs = 2
num_prefetch_proc = 1
/
&grid_nml
dynamics_grid_filename = ${dynamics_grid_filename}
dynamics_parent_grid_id = ${dynamics_parent_grid_id}
l_limited_area = .TRUE.
lfeedback = .FALSE. ! Daniel war TRUE
start_time = 0. , 60. , 120.
/
&initicon_nml
init_mode = 4 !4=cosmo, 2=ifs, 3=combined
lread_ana = .FALSE.
ifs2icon_filename = '${GRIDFOLDER}/iefff00000000.nc'
dwdfg_filename = '${GRIDFOLDER}/iefff00000000.nc'
/
&io_nml
itype_pres_msl = 5 ! IFS method with bug fix for self-consistency between SLP and geopotential
itype_rh = 1 ! RH w.r.t. water
restart_file_type = 5 ! 4: netcdf2, 5: netcdf4
dt_checkpoint = ${dt_checkpoint}
restart_write_mode = "joint procs multifile"
/
&limarea_nml
itype_latbc = 1
dtime_latbc = 3600
latbc_path = '${GRIDFOLDER}'
latbc_filename = 'iefff_<y><m><d><h>_lbc.nc'
latbc_boundary_grid = '${GRIDFOLDER}/lateral_boundary.grid.nc' ! Grid file defining the lateral boundary
init_latbc_from_fg = .TRUE. !Otherwise with this version of ICON you get the Internal Error in mtime
/
&run_nml
num_lev = 90
lvert_nest = .FALSE.
dtime = ${dtime} ! timestep in seconds
ldynamics = .TRUE. ! dynamics
ltransport = .TRUE.
iforcing = 3 ! NWP forcing
ltestcase = .FALSE. ! FALSE: run with real data
msg_level = 5 ! default: 5, much more: 20
ltimer = .TRUE.
timers_level = 10
activate_sync_timers = .FALSE.
output = 'nml'
check_uuid_gracefully = .TRUE.
/
&nwp_phy_nml
inwp_gscp = 2 ! 1: default, 2: graupel=NARVAL, 4: for two-moment=HDCP2
inwp_convection = 0
inwp_radiation = 1
inwp_cldcover = 1, 5, 5 ! 0: no cld, 1: new diagnostic, 3: COSMO, 5: grid scale
inwp_turb = 1, 5, 5 ! 1/10: Raschendorfer, 2: GME, 3: EDMF-DUALM (ntracer+1,ntiles=8)
inwp_satad = 1 != NARVAL
inwp_sso = 0 != NARVAL
inwp_gwd = 0 != NARVAL
inwp_surface = 1 != NARVAL
itype_z0 = 2 != NARVAL !1: default, 2: turn off SSO part of z0
dt_rad = 720.
latm_above_top = .FALSE. != NARVAL ! maybe should be set to true? (=take into account atmosphere above model top for radiation computation)
efdt_min_raylfric = 7200. != NARVAL
lrtm_filename = 'rrtmg_lw.nc'
cldopt_filename = 'rrtm_cldopt.nc'
/
&turbdiff_nml !!! Daniel Dynmod setup
tkhmin = 0.75 !=MBOld=NARVAL
tkmmin = 0.75 !=MBOld=NARVAL
tkmmin_strat = 4 ! NA in MBOld
pat_len = 750. !=MBOld=NARVAL
c_diff = 0.2 !=MBOld=NARVAL
rat_sea = 7.0 !=8.0 in MBOld
ltkesso = .TRUE.!=MBOld=NARVAL
frcsmot = 0.2 !=MBOld! these 2 switches together apply vertical smoothing of the TKE source terms
imode_frcsmot = 2 !=MBOld! in the tropics (only), which reduces the moist bias in the tropical lower troposphere
! use horizontal shear production terms with 1/SQRT(Ri) scaling to prevent unwanted side effects:
itype_sher = 3 !=MBOld=NARVAL
ltkeshs = .TRUE.!=MBOld=NARVAL
a_hshr = 2.0 !=MBOld=NARVAL
alpha0 = 0.0123!=MBOld=NARVAL
alpha0_max = 0.0335!=MBOld=NARVAL
icldm_turb = 1 !=NA in MBOld !Mode of water cloud representation in turbulence
/
&lnd_nml
ntiles = 1
nlev_snow = 3
lmulti_snow = .FALSE.
itype_heatcond = 2
idiag_snowfrac = 20
lsnowtile = .FALSE. !! later on .TRUE. if GRIB encoding issues are solved
lseaice = .TRUE.
llake = .TRUE.
itype_lndtbl = 3 ! minimizes moist/cold bias in lower tropical troposphere
itype_root = 2
/
&radiation_nml
irad_o3 = 7
irad_aero = 6
albedo_type = 2 ! 1: default, 2: MODIS
/
&nonhydrostatic_nml
iadv_rhotheta = 2
ivctype = 2
itime_scheme = 4
exner_expol = 0.333
vwind_offctr = 0.25
damp_height = 25000.
rayleigh_coeff = 0.25
lhdiff_rcf = .TRUE.
divdamp_order = 24 ! (does not allow checkpointing/restarting earlier than 2.5 hours of integration)
divdamp_type = 32
divdamp_fac = 0.004
l_open_ubc = .FALSE.
igradp_method = 2
l_zdiffu_t = .TRUE.
thslp_zdiffu = 0.02
thhgtd_zdiffu = 125.
htop_moist_proc = 22500.
hbot_qvsubstep = 19000.
/
&sleve_nml
min_lay_thckn = 20.
max_lay_thckn = 400. ! maximum layer thickness below htop_thcknlimit
htop_thcknlimit = 14000. ! this implies that the upcoming COSMO-EU nest will have 60 levels
top_height = 30000. !=NARVAL
stretch_fac = 0.9
decay_scale_1 = 4000.
decay_scale_2 = 2500.
decay_exp = 1.2
flat_height = 16000.
/
&dynamics_nml
iequations = 3
idiv_method = 1
divavg_cntrwgt = 0.50
lcoriolis = .TRUE.
/
&transport_nml
ivadv_tracer = 3,3,3,3,3
itype_hlimit = 3,4,4,4,4
ihadv_tracer = 52,2,2,2,2
/
&diffusion_nml
lhdiff_vn = .TRUE. ! diffusion on the horizontal wind field
lhdiff_temp = .TRUE. ! diffusion on the temperature field
lhdiff_w = .TRUE. ! diffusion on the vertical wind field
hdiff_order = 5 ! order of nabla operator for diffusion
itype_vn_diffu = 1 ! reconstruction method used for Smagorinsky diffusion
itype_t_diffu = 2 ! discretization of temperature diffusion
hdiff_efdt_ratio = 24.0 ! ratio of e-folding time to time step
hdiff_smag_fac = 0.025 ! scaling factor for Smagorinsky diffusion
lsmag_3d = .TRUE. ! drieg: test
/
&interpol_nml
nudge_zone_width = 4 ! Daniel=4, Matt=8 ; Total width (in units of cell rows) for lateral boundary nudging zone.
! If < 0 the patch boundary_depth_index is used.
lsq_high_ord = 3 ! Daniel=3, Matt=2
rbf_vec_scale_c = 0.09, 0.03, 0.010
rbf_vec_scale_v = 0.21, 0.07, 0.025
rbf_vec_scale_e = 0.45, 0.15, 0.050
/
&gridref_nml
grf_intmethod_e = 5 ! Daniel=6, Matt=5
grf_intmethod_ct = 2 ! Daniel=2, Matt=1
grf_tracfbk = 2 ! Daniel=2, Matt=1
l_mass_consvcorr = .TRUE. ! Daniel=TRUE, Matt=NA(default=FALSE)
/
&extpar_nml
itopo = 1
fac_smooth_topo = 0.1 ! Daniel=einkommentiert , Matt=NA
hgtdiff_max_smooth_topo = 750. ! Daniel=einkommentiert , Matt=NA
n_iter_smooth_topo = 1,1,2
heightdiff_threshold = 1000., 1000., 750.
/
&les_nml ! Daniel=einkommentiert , Matt=NA
smag_constant = 0.3
isrfc_type = 1
ldiag_les_out = .FALSE.
les_metric = .TRUE.
/
&output_nml
filetype = 5 ! output format: 2=GRIB2, 4=NETCDFv2
dom = -1 ! write all domains
mode = 1 ! 1: forecast
output_time_unit = 1 ! 1: seconds
output_bounds = 0., 280800., 1800.
steps_per_file = 24
include_last = .TRUE.
output_filename = '${EXPNAME}-2D'
ml_varlist = 'tqv_dia','tqc_dia','tqi_dia','t_2m','rh_2m','sp_10m','u_10m','v_10m','shfl_s','lhfl_s','tot_prec','clct','pres_msl','cape_ml','cin_ml'
filename_format = "<output_filename>_DOM<physdom>_<levtype>_<jfile>"
output_grid = .TRUE.
/
&output_nml
filetype = 5 ! output format: 2=GRIB2, 4=NETCDFv2
dom = -1 ! write all domains
mode = 1 ! 1: forecast
output_time_unit = 1 ! 1: seconds
output_bounds = 0., 280800., 1800.
steps_per_file = 24
include_last = .TRUE.
output_filename = '${EXPNAME}-3D'
pl_varlist = 'u','v','w','rh','temp','clc','geopot','qv','qc','qr','qi','div'
p_levels = 100000, 97500, 95000, 92500, 90000, 87500, 85000, 80000, 75000, 70000, 65000, 60000, 50000, 40000, 35000, 30000, 25000, 20000, 15000, 10000, 5000
output_grid = .TRUE.
/
EOF
#-----------------------------------------------------------------------------
# start experiment
# Take the executable that is in the folder
cp -p $MODEL ./icon.exe
$START icon.exe
finish_status=`cat finish.status`
echo $finish_status
echo "============================"
echo "Script run successfully: $finish_status"
echo "============================"
#-----------------------------------------------------------------------------
#-----------------------------------------------------------------------------
# check if we have to restart, ie resubmit
#if [ $finish_status = "RESTART" ] ; then
# if [ $finish_status = "OK" ] ; then
# touch ${restartSemaphoreFilename} # create the file "isRestartRun.sem" exists. in the next iteration the script will restart
# echo "restart next experiment..."
# this_script="${RUNSCRIPTDIR}/${job_name}"
# echo 'this_script: ' "$this_script"
# cd ${RUNSCRIPTDIR}
# ${submit} $this_script
# fi
#-----------------------------------------------------------------------------
exit
#-----------------------------------------------------------------------------
At the beginning of the script we set some common directories:
ICON_BASE_PATH=/work/mh0731/m300382/icon-git # Where the binaries are
EXPDIR=/work/mh0731/m300382/test_nwp_new/exp/test # Where the output of the simulation will be stored
GRIDFOLDER="/work/mh0731/m300382/test_nwp_new" # Where the grids are located
grid_1=italy_grid_nested_1231m
grid_2=italy_grid_nested_616m
grid_3=italy_grid_nested_308m
ln -sf ${GRIDFOLDER}/${grid_1}.nc ${EXPDIR}/ # We link the grid files in our experiment directory
ln -sf ${GRIDFOLDER}/${grid_2}.nc ${EXPDIR}/
ln -sf ${GRIDFOLDER}/${grid_3}.nc ${EXPDIR}/
atmo_dyn_grids="${grid_1}.nc ${grid_2}.nc ${grid_3}.nc" # This is then used by ICON to define the grid
dynamics_parent_grid_id=" 0, 1, 2 "
###### EXT DATA (default filename of external parameter input file, default:”<path>extpar_<gridfile>”)
EXTPARFOLDER=${GRIDFOLDER}
ln -sf ${EXTPARFOLDER}/extpar_${grid_1}.nc ${EXPDIR}/
ln -sf ${EXTPARFOLDER}/extpar_${grid_2}.nc ${EXPDIR}/
ln -sf ${EXTPARFOLDER}/extpar_${grid_3}.nc ${EXPDIR}/
Then we define the main features of the simulation
start_date="2018-10-29T00:00:00Z"
end_date="2018-10-30T00:00:00Z"
dtime=10 # time step in seconds
nhours_restart=180 # restart is done after 180 hours (basically never in our case)
dt_restart=`expr ${nhours_restart} \* 3600 `
nhours_checkpoint=12 # a checkpoint (needed to restar the simulation afterwards) is written every 12 hours
dt_checkpoint=`expr ${nhours_checkpoint} \* 3600 `
model_equations=3 # equation system
restart=.FALSE. # this simulation is not restarted
What follow is a list of all the namelists that will be used by ICON to set up the simulation. You can add options or even add namelists that are not used. Always read the Namelist_overview.pdf file contained in the icon repository for more informations about all the parameters that you can change.
The most important are
¶llel_nml
num_io_procs = 2 ! Asynchronous output
num_prefetch_proc = 1 ! Asynchronous read-in of boundary conditions
/
&grid_nml
lfeedback = .FALSE. ! two-way feedback between the nests
start_time = 0. , 60. , 120. ! delayed initialization of the nests so that you don't need an initial condition for every domain
/
! All the physical parametrizations
&nwp_phy_nml
inwp_gscp = 2
inwp_convection = 0
inwp_radiation = 1
inwp_cldcover = 1 ! 0: no cld, 1: new diagnostic, 3: COSMO, 5: grid scale
inwp_turb = 5 ! 1/10: Raschendorfer, 2: GME, 3: EDMF-DUALM (ntracer+1,ntiles=8)
inwp_satad = 1 != NARVAL
inwp_surface = 1 != NARVAL
/
Some important things to note. First of all we used the delay initialization by specifying the parameter start_time. This means that we don't have to worry about providing an initial condition for every domain but just one for the outermost domain: after 60 seconds the inner domain will be initialized by interpolation and at 90 second the innermost domain will also be initialized. How cool is that?
Second, if you want different settings for the domain you can do e.g. inwp_gscp = 2, 4, 4. This will work for every setting in every namelist that support different domains configurations.
Third, we use an asynchronous I/O (num_io_procs and num_prefetch_pro>0): this is really important for high resolution as the model does not have to wait to finish writing output to proceed further in the integration. This speeds everything up.
Cool! Now you have a lot of data to analyse.
Caveats
Sometimes the simulation crashes due to interpolation problems, especially at high resolution. You can check whether this is the case if you write the output very frequently and especially when the domain become active. If you see NaN in the domain this is the problem. This can be fixed by specifying explicitly the values of the rbf parameter which should be smaller than defaults. Here are some reference values
&interpol_nml ! 1km 600m 300m 150m
rbf_vec_scale_c = 0.09, 0.03, 0.010, 0.004
rbf_vec_scale_v = 0.21, 0.07, 0.025, 0.0025
rbf_vec_scale_e = 0.45, 0.15, 0.050, 0.0125
/
Restart files are also written asynchronously and split across multiple files (restart_write_mode = “joint procs multifile”) instead than in a single one. This speeds up the read-in of restart files if a simulation needs to be restarted.